Vol. 7 (2024), Article ID 246166, 13 pages
Research Article
DNA Binding and Antioxidant Activities of Novel Synthesized Fe(III), Ni(II) and Cu(II) Complexes Derived from Monodentate V-Shaped Schiff Bases
Ramadan M. Ramadan, Amr M. A. Naeem, Amir E. Aboelhasan, and Ayman A. Abdel Aziz
Chemistry Department, Faculty of Science, Ain Shams University, Cairo, Abbasia 11566, Egypt
Received 1 January 2024; Revised 23 February 2024; Accepted 25 February 2024; Published 9 June 2024
Ramadan M. Ramadan, Amr M. A. Naeem, Amir E. Aboelhasan, and Ayman A. Abdel Aziz, DNA Binding and Antioxidant Activities of Novel Synthesized Fe(III), Ni(II) and Cu(II) Complexes Derived from Monodentate V-Shaped Schiff Bases, Journal of Transition Metal Complexes, 7 (2024), art246166. doi:10.32371/jtmc/246166
A new series of six bioactive mononuclear transition metal complexes was prepared by reacting two V-shaped Schiff base ligands, namely, L1: 1-(4-((4-(dimethylamino)benzylidene)amino)phenyl)ethan-1-one and L2: 1-(4-((3,4-dimethoxybenzylidene)amino)phenyl)ethan-1-one, with a stochiometric ratio of 1:1 metal to ligand. Analytical measurements and spectroscopic analyses were used to characterize the complexes. The calf thymus-DNA (CT-DNA) binding characteristics of the free ligands and their complexes have been examined by electronic absorption titration, ethidium bromide-DNA displacement experiments and viscosity measurements. The results support that the ligands and their complexes bind to DNA via an intercalative binding mode, and the affinity for DNA is more strongly in the case of complexes when compared with ligands. Moreover, in vitro antioxidant scavenging studies have shown that the complexes exhibited potential antioxidant properties.
mononuclear complexes; V-shaped ligands; spectra; CT-DNA-binding; antioxidant activities
Interest in synthesis new bioactive Schiff bases as well as their metal complexes was accompanied with the development of the fields of bioinorganic and medicinal chemistry because of their versatile applications across many biological fields [1]. Many Schiff bases and their corresponding transition metal complexes were found to be considered as antimicrobial [2], antibacterial [3], antitubercular [4], antitumor [5], anticancer [6], antiviral [7], anti-inflammatory [8], antioxidant [9], cytotoxic [10], DNA binding and DNA cleavage [11] and antidiabetic activities [12]. Among those metal complexes, the derivatives built from the coordination of V-shaped Schiff base ligands showed interesting potential applications according to their pharmacological activity, mechanism of transformation, inhibitory properties and tautomerization reactions in biological systems [13,14,15,16]. Hazardous chemicals and carcinogens have the capacity to adhere to DNA strands, potentially disrupting their function as receptors within cells. This interference can result in defects during DNA replication, consequently fostering the growth of tumors. Consequently, DNA damage manifests within cancerous and tumor cells, leading to a cessation in cellular proliferation, which marks a successful phase in cancer recovery [17]. The study of interaction of metal complexes with nucleic acid has gained prominent research in the bioinorganic chemistry field [18,19]. From this point of view, it is especially essential to molecularly designing new complexes, which could be applied to the medicinal field. Particularly, the appropriateness and importance of inorganic molecules [20] intensified extensive research in drug discovery especially those for cancer chemotherapy treatments.
In continuation to our previous work [21,22,23,24],
here, we describe the synthesis and characterization of six new Cu2+, Fe3+ and Ni2+ metal complexes incorporated with two V-shaped Schiff base ligands, L1 and L2. The effects of substituents in the Schiff base ligands on their DNA interactions and antioxidant activities were also investigated.
Chemicals and solvents used in this study were of reagent grade type and were used without further purification. 3′-aminoacetophenone, 4-(dimethylamino)benzaldehyde, 3,4-dimethoxybenzaldehyde, CuCl2·2H2O, FeCl3, NiCl2·6H2O, absolute ethanol, calf thymus-DNA (CT-DNA) and ethidium bromide (EtBr) were obtained from Sigma-Aldrich. Experiments involving the interaction of complexes with the CT-DNA were conducted using a buffer of doubly distilled water containing 5 mM Tris and 50 mM NaCl, adjusted to a pH of 7.2 with HCl. A UV absorbance at 260 nm and 280 nm for a CT-DNA solution having a ratio around 1.8–1.9:1 indicated the significant absence of protein [25]. The concentration per nucleotide of CT-DNA was spectrophotometrically estimated by utilizing an extinction coefficient of 6600 M−1 cm−1 at 260 nm [26]. EtBr, EDTA and safranin were obtained from Sigma-Aldrich. Stock solutions of the complexes with concentration 2×10−3 M 5% DMSO were freshly prepared.
Micro analyses (C, H, N) were carried out by using Vario EL M Germany. Chloride contents in the complexes were estimated using the conventional gravimetric technique [27]. The progress of reactions was monitored using thin-layer chromatography sheets packed by silica gel of the type Merck60F254 and screened by a UV lamp. Fourier transform-infrared (KBr pellets) measurements were registered on a Unicam-Mattson 1000 FT-IR covering the range of 4000–400 cm−1. Mass spectra were conducted on a VG ZAB-HS (FAB) instrument and electrospray mass spectra (ESI-MS) were measured on a LQC system (Finnigan MAT, USA) using the CH3OH mobile phase. Magnetic susceptibilities of the reported solid complexes (Gouy’s method) were evaluated at room temperature using Sherwood Scientific Magnetic Balance. 1,1-Diphenyl-2-picrylhydrazyl (DPPH)-2,2-diphenyl-1-picrylhydrazyl (DPPH) was used as a calibrant. The magnetic moments were then estimated by using the relation
|
|
|
Electron spin resonance (ESR) data of the powdered copper complexes were executed at room temperature using an X-band EMX spectrometer (Bruker, Germany) and the DPPH acted as a reference. Molar conductivity measurements were done in DMSO solutions of 1×10−3 M (25 °C) using YSE conductance meter model 32. Emission measurements were achieved on a Jenway 6270 Fluorimeter; a Pulsed Xenon Lamp was operated as an excitation source. The biological activities findings (DNA binding interactions and antioxidant activities) were carried out using UV-Vis spectroscopy (25 °C in a DMSO and concentration of 1×10−5 M) utilized by Shimadzu UV-Vis 1800 spectrophotometer.
The Schiff base ligands (L1 and L2) were synthesized following the previously reported procedures (Scheme 1) [28]. A (1:1) mixture of 4′-aminoacetophenone and benzaldehydes derivatives (4-(dimethylamino)benzaldehyde and 3,4-dimethoxybenzaldehyde) was refluxed at 80 °C with stirring in ethanol for 30 min. The insoluble yellow crude was filtered and washed several times by warm ethanol. The obtained product was dried under vacuum.
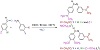
Scheme 1: Synthetic routes of the L1 and L2 Schiff bases.
A general procedure was employed for the synthesis of the complexes as follows: A 0.02 mol of the ligand in hot EtOH solution (20 mL) was added to 20 mL ethanol-water (1:1) solution (0.01 mol) of the appropriate metal chloride Ni(II), Cu(II) and Fe(III). The mixture was stirred under reflux for 2 h. The complexes were rapidly precipitated on cooling whereas the Ni(II) complex of L2 was prelimited by heating to reduce the volume of solution to about one-third the volume and leaving to precipitate. The obtained crude was filtered, washed several times by aqueous ethanol and then recrystallized from hot EtOH. The obtained product was left to dry in vacuo.
All the measurements including the interactions of the complexes with the CT-DNA were performed in DMSO solutions containing Tris-HCl buffer (20 mM HCl and 20 mM NaCl at pH 7.2). Stock solutions of CT-DNA (4 mg/mL) were prepared prior to the measurements using Tris-HCl buffer with gentle stirring until homogenous and then stored at −4 °C. Also, stock solutions of the complexes (1 × 10−2 M) were prepared using the same buffer. The purities of CT-DNA solutions were assured for the absence of the free of protein [29]. The DNA concentration per nucleotide (NP) was measured at 260 nm using the reported molar extinction coefficient (6,600 L.mol−1.cm−1) of DNA [30].
Electronic absorption titrations were done to determine the binding affinities between the DNA and the reported compounds. In the experiments, a fixed amount of the compound (5.0 μM) was titrated by raising the concentration of the CT-DNA from 0.0 mM to 90.0 mM. In addition, 3.0 mL solutions of the blank buffer and the complex sample (5.0 μM) were placed into two one cm cells, respectively. An aliquot (2.0 mL) of CT-DNA solution was then added to both the compound and blank solutions. The tested compound-DNA solutions were allowed to be incubated for 5 min. Each sample was scanned in the range of 200–500 nm to get the absorption spectrum. The quantitative DNA-binding affinity of the tested compound was estimated by computing the intrinsic binding constants (Kb) using the following modified Wolfe–Shimmer relation [31]:
|
|
where [compound] is the concentration of the tested sample; εa and εf were the molar extinction coefficient for the free CT-DNA and each addition of the compound to the CT-DNA, respectively. εb was the molar extinction coefficient of the CT-DNA when fully bound to the complex. The graph of
[compound]/(εa − εf) versus [compound] afforded a slope of 1/(εb − εf) and an intercept Y = 1/Kb(εb − εf). The constant Kb was the proportion of the slope to intercept.
Interactions of the ligands and their related metal derivatives with the CT-DNA were further explored using EtBr, a specific marker, and monitored by fluorescence spectroscopy as reported in the literature [32]. The competitive binding measurements were performed in buffer keeping the ratio [DNA]/[EtBr] = 1.1 with the variation of concentrations of complexes. In the fluorescence cell charged by 2.0 mL Tris-HCl buffer, 50.0 μL of 5.0 μM of the CT-DNA and 50.0 μL of 5.0 μM aqueous EtBr were added and permitted to equilibrate for 15 min. The fluorescence spectra of EtBr-DNA solutions were measured at 596 nm using an excitation wavelength of 510 nm where maximum quantum yield for EtBr was fulfilled. The effect of each complex on the emission intensity of the interacted EtBr-DNA adduct was then studied by increasing the concentrations of ingredients (0.0–50.0 μM). The spectra were analyzed to evaluate the KSV (Stern–Volmer quenching constant). This constant is a measure for the effectiveness of the complex as a quencher reagent. The quenching constant was estimated by using the classical Stern–Volmer equation [33]:
|
|
F and F0 were the fluorescence intensities at 596 nm in the presence and absence of the tested complex.
Viscosity measurements were executed on an Ubbelodhe viscometer in a water-bath maintained at 25.00 ± 0.01 °C. They were performed for a tested compound by introducing it into the DNA solution (50 mM, bps) present in the viscometer. Results were demonstrated as (η/η0) versus the compound/DNA ratio. η and η0 were the viscosity of DNA in the presence of the compound corrected from the solvent effect and the viscosity of DNA alone, respectively.
Relative viscosity for DNA in either the absence or presence of the compound was computed using the following relation:
t and t0 were the observed flow time of the DNA containing solution and the flow time of buffer, respectively [34].
Free radical scavenging activities of the reported derivatives were investigated in vitro by examining the capability of tested compounds to undergo scavenging of generated hydroxyl radicals in aqueous media via a Fenton type reaction [35]. Three mL portions of reaction mixture included 1.0 mL (100 mmol, pH = 7.4) of phosphate buffer, 1.0 mL (0.1 mmol) of aqueous safranin, 1.0 mL of aqueous EDTA-Fe(II) mmol, 1 mL (3%) of aqueous H2O2 and a set of quantitative micro additives of solutions of the tested compound. The reaction mixture was incubated at 37 °C in a water-bath for half an hour. Absorbance was measured at 520 nm during which the solvent effect was corrected throughout. The strong antioxidant ascorbic acid was used as a positive control. All tests were performed in triplicate and expressed as a mean and a standard deviation (SD) [36]. The percentage of OH scavenging was estimated using the following equation [37]:
|
|
AS, AB and AC were the absorbance of the sample in presence of the tested compound, the absorbance of blank in absence of the tested compound and the absorbance in absence of the tested compound and EDTA–Fe(II), respectively. The molar concentration of the tested compound which caused a 50% inhibition or scavenging effect on OH∗ radicals was denoted as IC50 value.
The energetically optimized structures of the two reported V-shaped Schiff bases were performed using density function theory as implemented in Gaussian 09w software. The DFT (B3LYP) method with the basis set 6-31G (d,p) and double zeta plus polarization for the ligand atoms was adopted.
All the reported Schiff base ligands and their complexes (1)–(6) are found to be air stable for extended periods. The analytical analyses and some physical characteristics of the reported compounds are tabulated in Tables 1 and 2. The complexes are soluble in hot ethanol and conventional polar organic solvents. The microanalytical analysis data showed that the monodentate Schiff base ligands L1 and L2 form 1:2 (M: L1–2) stoichiometric complexes with the Fe(III), Ni(II) and Cu(II) metal ions. The synthesized complexes were found to have a general formula [M(L1–2)2ClX]·yH2O, x = 2–3 and y = 0–2. The involvement of the chloride ions in coordination to metal ions was further concluded from the conductivity measurements. The molar conductivities of complexes in DMSO solution (1×10−3 M) were within the range of 5.8–13.3, which reflected the non-electrolyte characteristics [38]. Figures 1 and 2 depict the geometry optimization process for the synthesized ligands, which confirmed their V-shaped structure arrangements. The Schiff base ligands adopted a S2 point group symmetry as indicated from the final molecular structure optimization.

Table 1: Some physical parameters of the ligands L1, L2 and their complexes.

Table 2: Elemental analysis and magnetic moment of the ligands L1, L2 and their complexes.
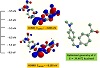
Figure 1: The geometry optimization, LOMO and HUMO orbitals for L1.
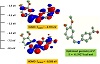
Figure 2: The geometry optimization, LOMO and HUMO orbitals for L2.
The IR spectra of the mononuclear complexes (Figures S1–S8) were compared with their parent ligands (L1 and L2). The most characteristic bands detected in the infrared spectra and their assignments are depicted in Table 3, showing that the vibrations of the ν(C⚌N) groups were shifted to higher frequencies due to the coordination of the nitrogen atom of the azomethine group [39]. The IR spectra of the ligands L1 and L2 showed that no peak was found for aldehyde carbonyl group stretching, with the appearance of a strong vibration band at 1,680 cm−1 and 1,673 cm−1, respectively, which are characteristic of the stretching vibration of the ν(C⚌N) azomethine group [40]. Upon coordination of the Schiff bases with metal ions, the azomethine, ν(C⚌N), vibration was shifted to the range of 1,677–1,686 cm−1, indicating the participation of the nitrogen atom of the Schiff base to the metal ion [41]. Moreover, the IR spectra of the complexes demonstrated new weak bands in the range of 411–429 cm−1 that can be due to the formation of M−N bonds. Also, the IR spectra of (4) and (6) showed broad bands at 3,444 cm−1 and 3,443 cm−1, respectively, corresponding to the OH groups of the coordinated water molecules [42].

Table 3: IR spectra data of the Schiff bases ligands and their metal complexes (1)–(6).
The complex behavior of transition metal complexes of the V-shaped Schiff bases was studied by electrospray ionization (ESI) mass spectrometry. The ES mass spectra of complexes are depicted in Figures S9–S14. The mass spectra of the compounds were in good agreement with the proposed structures. Positive ion electrospray (ES) mass spectra for the mononuclear complexes showed peaks related to fragments at m/z = 623.9 (77%; Fe complex (1)), 662.3 (41%; Ni complex (2)), 596.2 (79%; Cu complex (3)), 729.4 (80%; Fe complex (4)), 696.3 (52%; Ni complex (5)) and 702.1 (83%; Cu complex (6)), which were corresponding to [P−2Cl−2H2O]+, [P−2H2O]+, [P−2Cl]+, [P]+, [P]+ and [P−1]+, respectively. The occurrence of these fragments concluded that the ligand coordinated to the metal center.
Ni(II) complexes of the two Schiff bases, (2) and (5), were diamagnetic due to the square planar geometry adopted by the Ni(II) center [43]. The Cu(II) complexes showed magnetic susceptibility values as expected for a d9 transition metal center. The effective magnetic moment (μeff) values of the Cu(II) ions in the complexes (3) and (6) were found to be 1.84 BM and 1.91 BM, respectively, which was consistent with the expected spin-only magnetic moment of S = , d9 copper(II) system and characteristic for the square planar geometry of the Cu(II) complexes [44]. These values were slightly higher than the spin only value of 1.73 BM due to the presence of a low-lying excited term, which was able to mix some of its orbital angular momentum with the spin angular momentum of the ground state [45,46]. The room temperature magnetic moment of the Fe(III) complexes is of 5.21 BM for complex L1−Fe (1) and 4.98 BM for L2−Fe (3) characteristic of five unpaired electrons. The measured magnetic moment values of the complexes were observed to be specific for trigonal bipyramid geometry according to the literature [47,48].
The UV-Vis spectra of the V-shaped ligands (L1–2) and their metal derivatives (1)–(6) in DMSO solutions (1×10−5 M) were recorded (Table 4). The spectra of the Schiff bases showed two bands in the UV region ascertained to the π–π∗ transitions of the aromatic rings along with bands in the visible range corresponding to the n–π∗ intra-ligand transitions with the involvement of C⚌N group [49]. The electronic spectra of the complexes displayed shifts in the ligand bands accompanied by additional bands in the higher wavelength region signifying metal-ligand coordination and complex formation. The Ni(II) complex (5) exhibited an absorption band in the visible region at 431 nm. The d−d band in Ni(II) complex was assigned to 1A2g ← 1A1g transition, which is characteristic of the Ni(II) ion in a square planar environment [50]. In case of Cu(II) complex (6), the visible spectra contained d–d broad and unsymmetrical band, assigned to a 2B2g ← 2B1g transition, that a characteristic of the Cu(II) square planar complexes [50]. Interestingly, the other Cu(II) complex (3) did not show such transition; instead it exhibited a non-ligand band at 368 nm, and in view of its very high intensity, it was assigned to charge transfer arising from the transfer of charge from the ligand to the metal center, most probably, from the azomethine nitrogen atoms [51]. In the case of the Fe(III) complex (4), the lowest energy absorption band at 439 nm was assigned as ligand-to-metal charge transfer bands (LMCT) [52].

Table 4: The UV-Vis data and their assignments for L1, L2 and their complexes.
EPR spectra studies of Cu(II) complexes gave important information about the allocation of the unpaired electron and hence on the nature of bonding between metal ion and ligands. The EPR spectra of the copper complexes (Figures S15 and S16) provided valuable data for studying the Cu(II) ion environmental arrangements. The trend in the observed g values of complex (3) at room temperature was g∥ (2.19263) > g⊥ (2.07865) > (2.0023). On the other hand, the EPR spectrum of the copper(II) complex (6) exhibited an anisotropic spectrum with g∥ = 2.11933, g⊥ = 2.04922 and giso = 2.0726 computed from the expression giso = (g∥ + 2g⊥)/3. This trend in both complexes provided evidence about the localization of the unpaired electron in the dx2−y2 orbital. Additionally, the g value < 2.3 indicated the covalent characteristics of the Cu(II)−L bond in complex [53]. The axial symmetry parameter G is defined as
|
|
It is well known [54] that if G < 4, a significant exchange coupling is present between Cu(II) ions and causes broadening the g component, with splitting the g component and reflecting the coupling with Cu. However, the ESR spectra of the present Cu(II) complexes are similar in nature to those of the mononuclear complexes [55]. This indicated that the two paramagnetic centers of the complexes were equivalent and there was no super exchange interaction between the two metal centers [56]. Therefore, the g∥ and g⊥ values for both complexes were in good harmony with a dx2−y2 Cu2+ ground state and the d9 configuration would be (eg)4 (b2g)2 (a1g)2 (b1g)1 [57]. The greater value of g∥ than g⊥ in complex (6) indicated a tetragonal distortion with an inverse axial symmetry [58]. In addition, the g∥ amount in the copper(II) complexes is a parameter sensitive to the covalent property of the metal-ligand bond. For a covalent character, g∥ < 2.3 [59]. The g∥ value for the present Cu(II) complex (6) showed considerable mixed ionic-covalent bonding characteristics. The G factor suggested that the local tetragonal axes were only slightly misaligned and the exchange interactions between copper(II) centers in the solid state were negligible [60].
Based on the spectroscopic and physicochemical data, proposed structures for the reported complexes are represented in Figure 3.
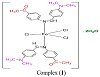
Figure 3: The proposed structures of the reported complexes.
Since the Human Genome Project announced the decoding of the human genome map, a large number of new specific therapeutic targets were provided [61]. Moreover, chemotherapy is one of the important treatments for cancer medication. Many compounds were invented as potential anticancer reagents, but only few of them have been used and registered as effective clinical drugs [62]. The mechanism of interaction of the anticancer drugs with DNA was endorseed as the most acceptable pathway. Thus, the DNA-interacting drugs could be categorized as (i) drugs that can form conventional covalent bond with DNA; (ii) drugs that execute non-covalent derivatives with DNA by either intercalation or groove-interaction; and (iii) drugs that have cleavage property of the DNA backbone [63]. Moreover, understanding the interactions of tiny molecules with specific sequences of DNA is a principal in any trial to control gene expression. In order to design new chemotherapeutic reagents, a major strategy is to evolve DNA binding substrates that can influence the crucial cellular processes like nucleic acid topology, replication, transcription and DNA repairing [64]. The knowledge about the affinity and binding modes of biologically active substrates versus the DNA helical may give some ideas about the ability to DNA damage, and consequently inhibiting the growth of the tumor cell [65]. Metal complex-DNA interactions display the influence, which the coordination geometry of the metal and the disposition of the ligands have on the binding activities. For instance, the square planar complexes allow deep insertion of the intercalator compared to those of octahedral or tetrahedral geometries [66]. In the present study, the evaluations of the electronic effect of phenyl ring substituents in the complexes of Schiff bases on the DNA binding behaviors were studied to provide rationalization for the difference in the binding properties of Schiff bases analogues having different substitution on the phenyl ring. In addition, hydrogen bonding formation between oxygen, fluorine and nitrogen is mostly known as proton acceptors in the biological systems [67].
Utilizing electronic absorption titration in DNA-binding studies stands as one of the most valuable methods for exploring both the potential binding mode and the binding affinity of compounds towards DNA. Electronic absorption spectroscopy serves as the predominant approach for investigating compound interactions with DNA. When a compound binds to DNA through intercalation interactions, it typically induces hypochromic and bathochromic effects. These effects are characterized by a robust π–π stacking interactions between the aromatic chromophore and the DNA base pairs [68]. Furthermore, the degree of hypochromism within the UV-Vis band aligns with the strength of the intercalative interaction.
The absorption spectra of the reported complexes in the absence and presence of the constant concentration of CT-DNA are given in Figures 4 and 5. The addition of increasing quantity of the metal complexes (1)–(6) to the CT-DNA solutions resulted in hypochromic shift in the UV-Vis spectra. These features are characteristic to the DNA intercalation binding and reflect involving a strong π–π∗ stacking interaction between the aromatic intercalative compound and the CT-DNA base pairs [69]. Furthermore, the substantial decreasing in the absorption intensity within the bands of the tested complexes in the presence of the double helical DNA commonly indicates the intercalation of molecules within DNA base pairs. This outcome arises from the potent stacking interaction between the aromatic chromophore and the DNA base pairs [69].

Figure 4: The absorption spectra of CT DNA (5 μM) in the absence (dashed line) and presence of 0.0–90.0 μM (solid line) of (a) Fe(III) complex (1), (b) Ni(II) complex (2) and (c) Cu(II) complex (3).

Figure 5: The absorption spectra of CT DNA (5 μM) in the absence (dashed line) and presence of 0.0–90.0 μM (solid line) of (d) Fe(III) complex (4), (e) Ni(II) complex (5) and (f) Cu(II) complex (6).
From the electronic absorption spectroscopy experimental data (Figure 6), the intrinsic binding constants Kb of the complexes (1)–(6) were found as 1.3404×105 M−1 (R2 = 0.9909), 2.2088×105 M−1 (R2 = 0.9993), 2.8670×105 M−1 (R2 = 0.09962), 1.4127×105 M−1 (R2 = 0.9951),
2.4097×105 M−1 (R2 = 0.9959) and 3.3390×105 M−1
(R2 = 0.9860), respectively. Comparing with the previously reported DNA intercalative complexes [70], our complexes can also bind to DNA via an intercalation mode. Thus, the order of binding affinities was Cu-complex (6) > Cu-complex (3) > Ni-complex (5) > Ni-complex (2) > Fe-complex (4) > Fe-complex (1). This order might be attributed to the fact that Cu(II) and Ni(II) complexes with large coplanar aromatic rings result in relatively close stacking between ligands and the DNA pairs [71]. Additionally, the above values of the intrinsic binding constant indicated that the binding affinities of the complexes having Schiff bases with methoxy substituents are stronger than those of the ligand bearing N-dimethyl substituents.

Figure 6: Plot of the ratio of [complexes (1)–(6)]/(εa–εf) vs. [complex].
The binding affinity of the complexes towards DNA is indirectly inferred from their strength to reduce fluorescence intensity in EtBr-DNA complexes, regardless of the specific binding mode. The planar cationic dye EtBr has the power to non-specifically intercalate into DNA to form soluble and intense fluorescent complexes and leads to strong fluorescence emission. This emission intensity is heightened due to the planar phenanthridinium ring stacking interactions between adjacent DNA base pairs. Upon the addition of CT-DNA to EtBr solution, the fluorescence intensity significantly increases. On the other hand, when the metal complex (quencher) is introduced to a treated DNA with EtBr, the fluorescence diminishes due to competition between the tested complex and EtBr for intercalating sites [72].
The reported complexes generally displayed no luminescence at room temperature either in organic solvents or in the presence of CT-DNA. The emission spectra of EtBr-CT-DNA adduct during titration with different concentration of metal complexes were monitored in the range of 525 nm to 725 nm (Figures 7 and 8). The spectra showed that the successive addition of the metal complex to EtBr-DNA adduct caused a gradual decrease of the fluorescence intensity. This decrease in the fluorescence intensity reflected the competition of the complexes with EtBr for DNA-binding, since EtBr intercalates DNA through binding interactions with its minor grooves [73]. The Stern–Volmer plots of EtBr-DNA showed that the quenching of EtBr-DNA adduct by the reported complexes followed a linear relationship, which is consistent with their intercalation (Figure 9). The Ksv values for the complex were 2.056×104 M−1 (R2 = 0.965), 3.156×104 M−1
(R2 = 0.9908), 4.174×104 M−1 (R2 = 0.99034), 2.361×104 M−1 (R2 = 0.9951), 3.642×104 M−1 (R2 = 0.9860) and 4.566×104 M−1 (R2 = 0.9907), for complexes (1)–(6), respectively. The data obtained were consistent with those obtained from the electronic absorption study (vide supra). The phenomena suggested that the complexes can compete for DNA-binding sites with EtBr and displace EtBr from the EtBr-DNA system. This behavior is usually characteristic of the intercalative interaction of compounds with DNA [74].

Figure 7: Emission spectra of EtBr-CT-DNA adduct in the absence (dashed line) and presence of (a) Fe(III) complex (1), (b)
Ni(II) complex (2) and (c) Cu(II) complex (3). The arrow displays the decrease in the intensity of EtBr-DNA upon increasing the concentration of the complex.

Figure 8: Emission spectra of EtBr-CT-DNA adduct in the absence (dashed line) and presence of (d) Fe(III) complex (4), (e)
Ni(II) complex (5) and (f) Cu(II) complex (6). The arrow displays the decrease in the intensity of EtBr-DNA upon increasing the concentration of the complex.

Figure 9: Plot of F/F0 versus [complexes (1)–(3)].
Viscosity measurements serve as a sensitive gauge for DNA length alterations and are considered crucial in determining binding modes. To elucidate the interaction modes between the studied complexes and CT-DNA, viscosity titration measurements were conducted. In the classical intercalative mode, there is a notable rise in DNA viscosity owing to an expansion in the separation of base pairs at intercalation sites, consequently increasing the overall DNA length. Conversely, complexes that exclusively bind to DNA grooves through partial and/or non-classical intercalation generally result in either minimal positive, negative, or no changes in DNA solution viscosity under similar conditions [75].
The effects of complexes on the viscosities of CT-DNA are shown in Figure 10. As the ratios of the investigated complexes to CT-DNA increase, the relative viscosities of CT-DNA increase steadily, indicating that there exist intercalations between the investigated metal complexes with CT-DNA helix. In addition, the relative viscosities of DNA increase with the order of Cu(II) complex > Ni(II) complexes > Fe(III) complexes. These orders suggest the extents of the unwinding and lengthening of DNA helix by compounds and the affinities of compounds binding to DNA, which may be due to the key roles of substituent effects and the larger coplanar structures of Cu(II) and Ni(II) complexes than those of Fe(III) complexes.

Figure 10: Effects of increasing the amounts of the complexes (1)–(3) on the relative specific viscosity of CT-DNA.
Since the tested complexes exhibit reasonable DNA-binding affinity, it is considered worthwhile to investigate other potential aspects, such as their antioxidant activity. The ability of tested compounds to scavenge hydroxyl radicals was compared with that of the well-known natural antioxidant vitamin C. The scavenging activities of the ligands and their complexes, respectively, with respect to different concentrations are represented graphically in Figure 11. The results marked that the HO scavenging of metal complexes is higher than those of their ligands, possibly in that the larger conjugated metal complexes can react with HO to form larger stable macromolecular radicals than ligands [76]. Moreover, it was found that the IC50 values of the samples ranged from 10.81 μg/mL to 34.13 μg/mL. Based on the comparison of the IC50 values (Figure 12) of the samples, they ranked in the following order: L2 > L1 > (5) > (2) > (6) > (3) > (4) > (1). The lower IC50 values observed in antioxidant assays demonstrate that the investigated compounds have a strong potential relative to the natural antioxidant ascorbic acid, allowing using them as scavengers to eliminate the radicals.

Figure 11: Scavenging effects of (a) L1 and its complexes (1)–(3), (b) L2 and its complexes (4)–(6) on HO∗ radical.

Figure 12: HO∗ radical scavenging activity of the Schiff base ligands and their complexes (1)–(6) in terms of IC50.
In summary, we have successfully synthesized six new complexes using two ligands. These complexes were thoroughly characterized, and it was determined that the iron complexes exhibited a trigonal bipyramidal geometry, while both the nickel and copper complexes displayed a square planar geometry. Through DNA binding studies, we discovered that the strength of interaction depended on the strength of hydrogen bonding. The order of strength was found to be Cu-complex (6) > Cu-complex (3) > Ni-complex (5) > Ni-complex (2) > Fe-complex (4) > Fe-complex (1). Furthermore, an antioxidant study was conducted, revealing that the antioxidant activity of the complexes was influenced by their ability to donate electrons. The order of electron donation affinity was observed to be Fe-complex (1) > Fe-complex (4) > Cu-complex (3) > Cu-complex (6) > Ni-complex (2) > Ni-complex (5). Based on the data obtained, it is evident that they hold potential as promising substances for use in chemotherapy.
The authors declare that they have no conflict of interest.
- M. N. Uddin, S. S. Ahmed, and S. M. R. Alam, Review: Biomedical applications of Schiff base metal complexes, J Coord Chem, 73 (2020), 3109–3149.
- R. Kumar, A. A. Singh, U. Kumar, et al., Recent advances in synthesis of heterocyclic Schiff base transition metal complexes and their antimicrobial activities especially antibacterial and antifungal, J Mol Struct, 1294 (2023), 136346.
- O. A. El-Gammal, F. S. Mohamed, G. N. Rezk, and A. A. El-Bindary, Synthesis, characterization, catalytic, DNA binding and antibacterial activities of Co(II), Ni(II) and Cu(II) complexes with new Schiff base ligand, J Mol Liq, 326 (2021), 115223.
- R. Cordeiro and M. Kachroo, Synthesis and biological evaluation of anti-tubercular activity of Schiff bases of 2-amino thiazoles, Bioorg Med Chem Lett, 30 (2020), 127655.
- K. Venkateswarlu, N. Ganji, S. Daravath, et al., Crystal structure, DNA interactions, antioxidant and antitumor activity of thermally stable Cu(II), Ni(II) and Co(III) complexes of an N,O donor Schiff base ligand, Polyhedron, 171 (2019), 86–97.
- T. A. Alorini, A. N. Al-Hakimi, S. El-Sayed Saeed, E. H. L. Alhamzi, and A. E. A. E. Albadri, Synthesis, characterization, and anticancer activity of some metal complexes with a new Schiff base ligand, Arab J Chem, 15 (2022), 103559.
- S. M. Abd El-Hamid, S. A. Sadeek, S. F. Mohammed, F. M. Ahmed, and M. S. El-Gedamy, N2O2-chelate metal complexes with Schiff base ligand: Synthesis, characterisation and contribution as a promising antiviral agent against human cytomegalovirus, Appl Organomet Chem, 37 (2023), e6958.
- Q.-U.-A. Sandhu, M. Pervaiz, A. Majid, et al., Review: Schiff base metal complexes as anti-inflammatory agents, J Coord Chem, 76 (2023), 1094–1118.
- V. Sumalatha, S. Daravath, A. Rambabu, G. Ramesh, and Shivaraj, Antioxidant, antimicrobial, DNA binding and cleavage studies of novel Co(II), Ni(II) and Cu(II) complexes of N, O donor Schiff bases: Synthesis and spectral characterization, J Mol Struct, 1229 (2021), 129606.
- B. Gowdhami, Y. Manojkumar, R. T. V. Vimala, et al., Cytotoxic cobalt (III) Schiff base complexes: in vitro anti-proliferative, oxidative stress and gene expression studies in human breast and lung cancer cells, Biometals, 35 (2022), 67–85.
- M. Samuel and N. Raman, Comprehensive biological evaluation (DNA-binding, cleavage, and antimicrobial activity) of β-diketimine Schiff base ligands and their Cu(II) and Zn(II) complexes, J Coord Chem, 74 (2021), 2069–2091.
- H. R. Afzal, N. U. H. Khan, K. Sultana, et al., Schiff bases of pioglitazone provide better antidiabetic and potent antioxidant effect in a streptozotocin-nicotinamide-induced diabetic rodent model, ACS Omega, 6 (2021), 4470–4479.
- R. M. Ramadan, A. K. Abu Al-Nasr, and A. F. Noureldeen, Synthesis, spectroscopic studies, antimicrobial activities and antitumor of a new monodentate V-shaped Schiff base and its transition metal complexes, Spectrochim Acta A Mol Biomol Spectrosc, 132 (2014), 417–422.
- X. J. Shi, P. Y. Chen, M. Z. Wu, D. C. Cai, and L. Tian, Transition metal coordination complexes based on V-shaped bis-triazole ligand: syntheses, structures, and properties, J Coord Chem, 71 (2018), 1063–1072.
- H. Wu, Z. Yang, F. Wang, et al., V-shaped ligand 1,3-bis(1-ethylbenzimidazol-2-yl)-2-thiapropane and manganese(II), cobalt(II) and copper(II) complexes: Synthesis, crystal structure, DNA-binding properties and antioxidant activities, J Photochem Photobiol B, 148 (2015), 252–261.
- N. H. Nasaruddin, S. N. Ahmad, S. S. Sirat, et al., Structural characterization, theoretical and antibacterial study of a V-shaped, cyclohexane bridged ONNO Schiff base, J Mol Struct, 1248 (2022), 131489.
- B. J. Pages, J. Sakoff, J. Gilbert, et al., Combining the platinum(II) drug candidate kiteplatin with 1,10-phenanthroline analogues, Dalton Trans, 47 (2018), 2156–2163.
- A. Erxleben, Transition metal salen complexes in bioinorganic and medicinal chemistry, Inorg Chim Acta, 472 (2018), 40–57.
- M. Niu, M. Hong, G. Chang, X. Li, and Z. Li, A comparative study of cytotoxicity and interaction with DNA/protein of five transition metal complexes with Schiff base ligands, J Photochem Photobiol B, 148 (2015), 232–241.
- K. D. Mjos and C. Orvig, Metallodrugs in medicinal inorganic chemistry, Chem Rev, 114 (2014), 4540–4563.
- R. M. Ramadan, W. M. Elsheemy, N. S. Hassan, and A. A. Abdel Aziz, Synthesis, spectroscopic characterization, thermal behaviour, in vitro antimicrobial and anticancer activities of novel ruthenium tricarbonyl complexes containing monodentate V-shaped Schiff bases, Appl Organometal Chem, 32 (2018), e4180.
- R. M. Ramadan, S. M. El-Medani, A. Makhlouf, et al., Spectroscopic, density functional theory, nonlinear optical properties
and in vitro biological studies of Co(II), Ni(II), and Cu(II) complexes of hydrazide Schiff base derivatives, Appl Organomet Chem, 35 (2021), e6246.
- S. E. Mohamed, R. M. Ramadan, A. E. Aboelhasan, and A. A. Abdel Aziz, Design, synthesis, biomedical investigation, DFT calculation and molecular docking of novel Ru(II)-mixed ligand complexes, J Biomol Struct Dyn, 41 (2023), 1233–1252.
- A. A. Abdel Aziz, R. M. Ramadan, M. E. Sidqi, and M. A. Sayed, Structural characterisation of novel mononuclear Schiff base metal complexes, DFT calculations, molecular docking studies, free radical scavenging, DNA binding evaluation and cytotoxic activity, Appl Organomet Chem, 37 (2023), e6954.
- Y. Ni, D. Lin, and S. Kokot, Synchronous fluorescence, UV-visible spectrophotometric, and voltammetric studies of the competitive interaction of bis(1,10-phenanthroline)copper(II) complex and neutral red with DNA, Anal Biochem, 352 (2006), 231–242.
- R. Min, X. Hu, X. Yi, and S. Zhang, Synthesis, structure, DNA binding and cleavage activity of a new copper(II) complex of bispyridylpyrrolide, J Cent South Univ, 22 (2015), 1619–1625.
- G. H. Jeffery, J. Bassett, J. Mendham, and R. C. Denney, Vogel’s Textbook of Quantitative Chemical Analysis, John Wiley & Sons, New York, 5th ed., 1989.
- R. M. Ramadan, W. M. Elsheemy, N. S. Hassan, and A. A. Abdel Aziz, Synthesis, spectroscopic characterization, thermal behaviour, in vitro antimicrobial and anticancer activities of novel ruthenium tricarbonyl complexes containing monodentate V-shaped Schiff bases, Appl Organometal Chem, 32 (2018), e4180.
- E. Chalkidou, F. Perdih, I. Turel, D. P. Kessissoglou, and G. Psomas, Copper(II) complexes with antimicrobial drug flumequine: Structure and biological evaluation, J Inorg Biochem, 113 (2012), 55–65.
- S. Purtaş, M. Köse, F. Tümer, M. Tümer, A. Gölcü, and G. Ceyhan, A novel porphyrin derivative and its metal complexes: Electrochemical, photoluminescence, thermal, DNA-binding and superoxide dismutase activity studies, J Mol Struct, 1105 (2016), 293–307.
- S. Jahani, M. Khorasani-Motlagh, and M. Noroozifar, DNA interaction of europium(III) complex containing 2,2′-bipyridine and its antimicrobial activity, J Biomol Struct Dyn, 34 (2016), 612–624.
- A. G. Krishna, D. V. Kumar, B. M. Khan, S. K. Rawal, and K. N. Ganesh, Taxol-DNA interactions: fluorescence and CD studies of DNA groove binding properties of taxol, Biochim Biophys Acta, 1381 (1998), 104–112.
- R. Yousefi, R. Mohammadi, A. Taheri-Kafrani, et al., Study of the interaction between two newly synthesized cyclometallated platinum (II) complexes and human serum albumin: Spectroscopic characterization and docking simulation, J Lumin, 159 (2015), 139–146.
- R. Palchaudhuri and P. J. Hergenrother, DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action, Curr Opin Biotechnol, 18 (2007), 497–503.
- H. Wu, J. Yuan, G. Pan, et al., The V-shaped ligand bis(N-allylbenzimidazol-2-ylmethyl)benzylamine and its Ag(I) complex: synthesis, crystal structure, DNA-binding properties and antioxidation, J Photochem Photobiol B, 122 (2013), 37–44.
- Z. Guo, R. Xing, S. Liu, et al., The synthesis and antioxidant activity of the Schiff bases of chitosan and carboxymethyl chitosan, Bioorg Med Chem Lett, 15 (2005), 4600–4603.
- D. Huang, B. Ou, and R. L. Prior, The chemistry behind antioxidant capacity assays, J Agric Food Chem, 53 (2005), 1841–1856.
- I. Ali, W. A. Wani, and K. Saleem, Empirical formulae to molecular structures of metal complexes by molar conductance, Synth React Inorg Met-Org Nano-Met Chem, 43 (2013), 1162–1170.
- A. Bartyzel, Synthesis, thermal study and some properties of N2O4—donor Schiff base and its Mn(III), Co(II), Ni(II), Cu(II) and Zn(II) complexes, J Therm Anal Calorim, 127 (2017), 2133–2147.
- J. Sanmartín, M. R. Bermejo, A. M. García-Deibe, I. M. Rivas, and A. R. Fernández, Zinc and cadmium complexes with versatile hexadentate Schiff base ligands. The supramolecular self-assembly of a 3-D cage-like complex, Dalton Trans, 22 (2000), 4174–4181.
- S. Maikoo, L. M. K. Dingle, A. Chakraborty, B. Xulu, A. L. Edkins, and I. N. Booysen, Synthetic, characterization and cytotoxic studies of ruthenium complexes with Schiff bases encompassing biologically relevant moieties, Polyhedron, 184 (2020), 114569.
- N. Ribeiro, S. Roy, N. Butenko, et al., New Cu(II) complexes with pyrazolyl derived Schiff base ligands: Synthesis and biological evaluation, J Inorg Biochem, 174 (2017), 63–75.
- S. Basak, S. Sen, S. Mitra, C. Marschner, and W. S. Sheldrick, Square planar complexes of Cu(II) and Ni(II) with N2O donor set of two Schiff base ligands: synthesis and structural aspects, Struct Chem, 19 (2008), 115–121.
- K. A. Melha, Antimicrobial, spectral, magnetic and thermal studies of Cu(II), Ni(II), Co(II), UO2(VI) and Fe(III) complexes of the Schiff base derived from oxalylhydrazide, J Enzyme Inhib Med Chem, 23 (2008), 285–295.
- R. S. Joseyphus and M. S. Nair, Synthesis, characterization and
biological studies of some Co(II), Ni(II) and Cu(II) complexes derived from indole-3-carboxaldehyde and glycylglycine as Schiff base ligand, Arab J Chem, 3 (2010), 195–204.
- D. Moon, M. S. Lah, R. E. Del Sesto, and J. S. Miller, The effect of ligand charge on the coordination geometry of an Fe(III) ion: five- and six-coordinate Fe(III) complexes of tris(2-benzimidazolylmethyl)amine, Inorg Chem, 41 (2002), 4708–4714.
- D. H. Shi, N. Zhang, W. W. Liu, L. L. Gao, Q. Zhang, and Z. L. You, Synthesis, crystal structures, and biological activity of Cu(II), Mn(III), and Fe(III) complexes derived from N,N′-bis(4-methoxysalicylidene)ethylenediamine, Synth React Inorg Met-Org Nano-Met Chem, 42 (2012), 1177–1182.
- C. Krüger, P. Augustín, L. Dlháň, et al., Iron(III) complexes with pentadentate Schiff-base ligands: Influence of crystal packing change and pseudohalido coligand variations on spin crossover, Polyhedron, 87 (2015), 194–201.
- N. K. Chaudhary and P. Mishra, Metal complexes of a novel Schiff base based on penicillin: Characterization, molecular modeling, and antibacterial activity study, Bioinorg Chem Appl, 2017 (2017), 6927675.
- A. M. Mesubi, U. B. Eke, O. M. Oyeku, and S. O. Owalude, Spectral studies of transition metal complexes of dihydrobis(1-pyrazoyl)borate with some Lewis bases, J Taibah Univ Sci, 10 (2016), 70–79.
- A. Ahmed and R. Lal, Synthesis, characterization and electrochemical studies of copper(II) complexes derived from succinoyl- and adipoyldihydrazones, Arab J Chem, 10 (2017), S901–S908.
- A. Lugosan, S. R. Todtz, A. Alcázar, M. Zeller, J. J. Devery III, and W. T. Lee, Synthesis and characterization of trigonal bipyramidal FeIII complexes and their solution behavior, Polyhedron, 208 (2021), 115384.
- D. S. El-Sayed, E. M. Tawfik, A. F. Elhusseiny, and A. El-Dissouky, A perception into binary and ternary copper (II) complexes: synthesis, characterization, DFT modeling, antimicrobial activity, protein binding screen, and amino acid interaction, BMC Chem, 17 (2023), 55.
- K. Jagadesh Babu, S. Daravath, M. Swathi, D. Ayodhya, and Shivaraj, Synthesis, anticancer, antibacterial, antifungal, DNA interactions, ADMET, molecular docking, and antioxidant evaluation of novel Schiff base and their Co(II), Ni(II) and Cu(II) complexes, Results Chem, 6 (2023), 101121.
- A. Zülfikaroğlu, Ç. Y. Ataol, E. Çelikoğlu, U. Çelikoğlu, and Ö. İdil, New Cu(II), Co(III) and Ni(II) metal complexes based on ONO donor tridentate hydrazone: Synthesis, structural characterization, and investigation of some biological properties, J Mol Struct, 1199 (2020), 127012.
- S. Mandal, R. Sadhukhan, U. Ghosh, et al., Synthesis and characterization of two Cu(II) complexes with a new pyrazole-based Schiff base ligand: crystallography, DNA interaction and antimicrobial activity of Ni(II) and Cu(II) complexes, J Coord Chem, 69 (2016), 1618–1634.
- V. P. Singh, Synthesis, electronic and ESR spectral studies on copper(II) nitrate complexes with some acylhydrazines and hydrazones, Spectrochim Acta A Mol Biomol Spectrosc, 71 (2008), 17–22.
- M. S. Ghurab, O. A. El-Gammal, M. M. El-Gamil, and G. M. Abu El-Reash, Preparation, investigation, DFT, pH-metric and cyclic voltammetry of Cr(III), Fe(III), Co(II), Ni(II) and Cu(II) complexes derived from 2-(2-((2Z,3Z)-3-(hydroxyimino) butan-2-ylidene) hydrazineyl)-2-oxo-N-(pyridin-2-yl) acetamide (H3BYPA) and evaluation of their biological activity, J Mol Struct, 1272 (2023), 134156.
- N. V. Loginova, T. V. Koval’chuk, R. A. Zheldakova, et al., Copper (II) complexes of sterically hindered o-diphenol derivatives: synthesis, characterization and microbiological studies, Cent Eur J Chem, 4 (2006), 440–457.
- A. M. Ajlouni, Q. Abu-Salem, Z. A. Taha, A. K. Hijazi, and W. Al Momani, Synthesis, characterization, biological activities and luminescent properties of lanthanide complexes with [2-thiophenecarboxylic acid, 2-(2-pyridinylmethylene)hydrazide] Schiff bases ligand, J Rare Earths, 34 (2016), 986–993.
- J. C. García-Ramos, R. Galindo-Murillo, F. Cortés-Guzmán, and L. Ruiz-Azuara, Metal-based drug-DNA interactions, J Mex Chem Soc, 57 (2013), 245–259.
- R. Hajian, P. Hossaini, Z. Mehrayin, P. M. Woi, and N. Shams, DNA-binding studies of valrubicin as a chemotherapy drug using spectroscopy and electrochemical techniques, J Pharm Anal, 7 (2017), 176–180.
- A. Bora, S. K. Maiti, A. Singh, and P. Barman, Studies on the effect of remote substituents on the DNA binding activity of novel chiral Schiff bases, J Mol Struct, 1234 (2021), 130179.
- C. D. Andersson, B. K. Mishra, N. Forsgren, F. Ekström, and A. Linusson, Physical mechanisms governing substituent effects on arene-arene interactions in a protein milieu, J Phys Chem B, 124 (2020), 6529–6539.
- N. N. Rao, E. kishan, K. Gopichand, R. Nagaraju, A. M. Ganai, and P. V. Rao, Design, synthesis, spectral characterization, DNA binding, photo cleavage and antibacterial studies of transition metal complexes of benzothiazole Schiff base, Chem Data Collect, 27 (2020), 100368.
- B. J. Pages, D. L. Ang, E. P. Wright, and J. R. Aldrich-Wright, Metal complex interactions with DNA, Dalton Trans, 44 (2015), 3505–3526.
- B. G. de Oliveira and R. C. M. U. de Araújo, Hydrogen bonds and stacking interactions on the DNA structure: A topological view of quantum computing, in Advances in Quantum Theory, I. I. Cotaescu, ed., IntechOpen, Rijeka, 2012, ch. 6.
- C. S. Devi, B. Thulasiram, S. Satyanarayana, and P. Nagababu, Analytical techniques used to detect DNA binding modes of ruthenium(II) complexes with extended phenanthroline ring, J Fluoresc, 27 (2017), 2119–2130.
- B. Thulasiram, C. S. Devi, Y. P. Kumar, R. R. Aerva, S. Satyanarayana, and P. Nagababu, Correlation between molecular modelling and spectroscopic techniques in investigation with DNA binding interaction of ruthenium(II) complexes, J Fluoresc, 27 (2017), 587–594.
- H. Wu, G. Pan, Y. Bai, et al., Preparation, structure, DNA-binding properties, and antioxidant activities of a homodinuclear erbium(III) complex with a pentadentate Schiff base ligand, J Chem Res, 38 (2014), 211–217.
- W. X. Hong, F. Huang, T. Huan, et al., Comparative studies on DNA-binding and in vitro antitumor activity of enantiomeric ruthenium(II) complexes, J Inorg Biochem, 180 (2018), 54–60.
- K. Jana, S. Das, H. Puschmann, et al., Supramolecular self-assembly, DNA interaction, antibacterial and cell viability studies of Cu(II) and Ni(II) complexes derived from NNN donor Schiff base ligand, Inorganica Chim Acta, 487 (2019), 128–137.
- N. Deepika, Y. P. Kumar, C. Shobha Devi, P. V. Reddy, A. Srishailam, and S. Satyanarayana, Synthesis, characterization, and DNA binding, photocleavage, cytotoxicity, cellular uptake, apoptosis, and on-off light switching studies of Ru(II) mixed-ligand complexes containing 7-fluorodipyrido[3,2-a:2′,3′-c]phenazine, J Biol Inorg Chem, 18 (2013), 751–766.
- M. Flamme, E. Clarke, G. Gasser, and M. Hollenstein, Applications of ruthenium complexes covalently linked to nucleic acid derivatives, Molecules, 23 (2018), 1515.
- N. Kumar, R. Kaushal, and P. Awasthi, Non-covalent binding studies of transition metal complexes with DNA: A review, J Mol Struct, 1288 (2023), 135751.
- Ç. Aşilioğlu, S. Uzunboy, S. Demirci-Çekiç, and R. Apak, Colorimetric determination of sulfoxy radicals and sulfoxy radical scavenging-based antioxidant activity, ACS Omega, 8 (2023), 36764–36774.
Copyright © 2024 Ramadan M. Ramadan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.