Vol. 7 (2024), Article ID 246167, 11 pages
Research Article
Palladium(II) Chelates as Biologically Active Metallo-Drugs: Synthesis, Characterization, DNA Binding, Electrochemical and In Vitro Cytotoxic Studies
Radwa Hatem,2 Menna T. Moataz,2 Ayatallah Tarek,2 Philopateir Emil,2 Maryam Khaled,2 Lara Emad,2 and Azza A. Shoukry1
1Department of Chemistry, Faculty of Science, Cairo University, Giza, Egypt
2Biotechnology and Biomolecular Chemistry Program, Faculty of Science, Cairo University, Giza, Egypt
Received 2 January 2024; Revised 1 July 2024; Accepted 9 July 2024; Published 29 December 2024
Radwa Hatem, Menna T. Moataz, Ayatallah Tarek, Philopateir Emil, Maryam Khaled, Lara Emad, and Azza A. Shoukry, Palladium(II) Chelates as Biologically Active Metallo-Drugs: Synthesis, Characterization, DNA Binding, Electrochemical and In Vitro Cytotoxic Studies, Journal of Transition Metal Complexes, 7 (2024), art246167. doi:10.32371/jtmc/246167
With the purpose of studying the binding behavior of Pd(II) complexes with DNA as the main biological target, and their ability to penetrate reasonably into tumor cells and destroy their replication ability, in this study, four new complexes [Pd(TAB)(H2O)2]2+ (1), [Pd(en)(H2O)2]2+ (2), [Pd(TAB)meth] (3), [Pd(en)sar] (4) (where (TAB) is 3,4,3′,4′-tetraaminobiphenyl hydrochloride; (en) is ethylenediamine; (meth) is methionine; (sar) is sarcosine) have been synthesized and characterized by conventional physicochemical analysis. The binding properties of the complexes with CT-DNA were investigated by electronic absorption spectra. The intrinsic binding constants (Kb) calculated from UV-vis absorption studies were 8.36 × 103 M−1 and 4.25 × 103 M−1 for complexes (1) and (2), respectively. Thermal denaturation has been systematically studied by spectrophotometric method and the calculated ΔTm was nearly 5 °C for each complex. All the results suggest that the interaction modes between the complexes and CT-DNA were electrostatic and/or groove binding. The redox behavior of the two complexes was investigated by cyclic voltammetry. Both complexes, in the presence and absence of CT-DNA, show a quasi-reversible wave. The changes in E1/2, ΔE and Ipc/Ipa ascertain the interaction of complexes with CT-DNA. In addition, the antitumor activity of the complexes was tested on two cancer cell lines: the colon cancer cell line (HCT) and breast cancer cell line (MCF-7), as well as one normal cell line: the human normal melanocytes (HFB4). The results showed that complex (1) was a more potent antitumor agent than complex (2). The in vitro antimicrobial activity of the complexes was carried out using the disc diffusion method against different species of pathogenic bacteria and fungi. The activity data showed the all complexes are highly reactive in inhibiting the growth of the tested organisms.
Pd(II) chelates; calf thymus DNA, UV-vis spectroscopy; cyclic voltammetry, biological activity
The binding of metal complexes with DNA strands is of interest for both therapeutic and scientific reasons. Certain metal complexes have been proven to cleave the DNA
strand and can thus be employed as chemotherapeutic drugs [1]. Mixed-ligand complexes of 3D transition metals
have been discovered to be particularly beneficial because of their ability to bind DNA via a variety of interactions and to split the duplex by chemical, electrochemical, and photochemical reactivities [2,3].
Cis-platin and its analogues are currently one of the most effective chemotherapeutic drugs in use as first-line treatment for cancer [4]. Diamminedichloroplatinum(II), also known as cis-platin or cis-[PtCl2(NH3)2], has a well-established
effectiveness against ovarian and testicular cancer. It exerts its anticancer activity through interactions with DNA [5,6]. The cis-platin induced inhibition of DNA synthesis due to a change in the DNA template by binding with the complex. However, cis-platin has a limited range of activity and its use in clinical applications is limited due to side effects including neurotoxicity, nephrotoxicity, ototoxicity, vomiting, nausea and myelosuppression [7]. So, the dose given to patients has to be limited [8]. Studies proved that platinum complexes can bind and inactivate the thiol-containing enzymes of the renal causing serious side effects to the kidney [9]. In addition to the substantial side effects, therapeutic efficacy is also limited by intrinsic or treatment-induced resistant tumor cells. Alternative chemotherapeutic techniques have arisen as a consequence of these disadvantages. Alternative chemotherapeutic strategies have arisen as a result of these drawbacks. It is important to minimize side effects and improve the therapeutic effects of anticancer agents for the quality of life of patients.
Due to the analogy between the coordination chemistry of Pt(II) and Pd(II) complexes and the side effects of Pt(II) complexes, several labile Pd(II) complexes have been found to be useful as models for obtaining a reasonable picture of the thermodynamics of the reactions for closely related Pt(II) complexes [10]. Pd(II) analogues are useful model compounds based on the mechanistic research of the mechanism of action of Pt(II) anticancer medicines since they have the ability to exchange ligands 104–105 times faster than the corresponding Pt(II) analogues [11]. So, Pd(II) complexes do not maintain their structural integrity in biological fluids for long time until they reach the pharmacological particles. Previous studies have demonstrated that ligands have to be chosen properly, as they play an important role in modifying reactivity and lipophilicity, stabilizing certain oxidation states and imparting substitution inertness [12]. It has been found that Pd(II) complexes of different donor atom ligands have anti-inflammatory, antimicrobial (antimalarial, antitrypanosomal, antiamoebic) [13], antitumor (antiproliferation in multiple tumor cell lines including Pam212, HeLa, Vero) cell line [14]. Palladium complexes have lower kidney toxicity than cis-platin because the proteins in kidney tubules cannot replace the tightly bound Pd(II) chelate ligands with sulfhydryl groups [15,16].
The chemical structure of amine metal complex allows it to intercalate with DNA in a square-planar shape [17]. It has been proven that a bis-amino acids palladium(II) complex can selectively interact with biological components in the cell, particularly cell DNA, and display its biological functions [18]. Furthermore, a coordinated amino acid has the advantage of exerting specificity while remaining nontoxic when released inside the cell [15]. Several studies suggested that adding amino acids to Pd(II) complexes has an important role as they improve antiproliferative properties, reducing destructive structural stimulating effects regarding the carrier proteins and increased drug concentration in tumor cells [19]. In solutions, Pd(II) amine complexes have the ability to penetrate tumor cells, bind effectively to DNA and destroy the ability of tumor cells to replicate [10]. It was previously reported [20] that methionine enhances the efficacy of chemotherapeutic agents and increases the DNA methylation of genes while down-regulating gene expression [21]. Also, it has been proven that the sulfur in methionine strongly interacts with DNA and forms stable Pd-DNA adduct. Sarcosine is an amino acid found in a variety of living organisms as an amino acid metabolic intermediate and a component of peptides [22].
With the above in mind, in this work, we have synthesized two binary diaqua Pd(II) amine complexes viz (diaquatetraminobiphenylpalladium(II)) [Pd(TAB)(H2O)2]2+ (1) and (diaquaethylenediaminepalladium(II)) [Pd(en)(H2O)2]2+ (2), and two mixed-ligand amino acid complexes viz (tetraaminobiphenylmethionine palladium(II)) [Pd(TAB)(meth)] (3) and (ethylenediaminesarcosinepallidium(II)) [Pd(en)(sar)] (4).
The complexes were characterized by conventional physicochemical analysis as well as FTIR spectroscopy. The binding interaction with CT-DNA has been investigated with a variety of the techniques viz electronic absorption spectroscopy, thermal denaturation and cyclic voltammetry. Also, the complexes have been screened for their antimicrobial activities against some selected Gram-positive and Gram-negative bacteria.
Palladium(II) chloride was purchased from Acros organics. The ligands 3,4,3′,4′-tetraaminobiphenyl hydrochloride, ethylenediamine, sarcosine (N-methylglycine) and methionine were from Aldrich and Fluka. Calf thymus DNA was provided from Sigma. All solvents used were of the analar grade. All DNA-binding experiments were carried out in Tris-HCl buffer solution (50 mM NaCl, 5 mM Tris-HCl, pH 7.1). Tris-HCl buffer was prepared using deionized triple distilled water. Solutions of CT-DNA in buffer gave a ratio of UV-vis absorbance of 2.5:2.32 at 260 and 280 nm, respectively, indicating that the DNA was sufficiently free of protein [23]. The concentration of DNA was determined spectrophotometrically (ε260 = 6600 M−1 cm−1) [24]. Stock solution of DNA was stored at −200 °C. Concentrated stock solution of the palladium complexes (100 × 10−6 M) was prepared by dissolving an appropriate amount of the complex into 50 mL of deionized doubly distilled water and diluted suitably with Tris-HCl buffer to the required concentrations for all the experiments.
The CHNS analysis of the separated solid complexes was performed in the Microanalytical center, Cairo University. Absorption titration experiments were made using TB-85 thermobath Shimadzu model UV spectrophotometer. Cyclic voltammetry measurements were made on an EG&G PAR model 253 VersaStat potentiostat/galvanostat with electrochemical analysis software 273 using a three-electrode setup comprising a glassy carbon working, platinum wire auxiliary and a saturated Ag/AgCl (KCl) reference electrode. IR spectra were measured on 80486-pc FTIR Shimadzu spectrophotometer using KBr pellets.
Synthesis of [Pd(TAB)(H2O)2]2+ (1), [Pd(en)(H2O)2]2+ (2) complexes is as follows. Complexes [Pd(TAB)Cl2] and [Pd(en)Cl2] were prepared by heating PdCl2 (0.0324 g, 1.0 mM) and (0.0194 g, 1.0 mM) in 40 mL H2O, respectively, and KCl (0.0248 g, 2.0 mM equivalent) and (0.0163 g,
2.0 mM equivalent) in 10 mL H2O, respectively. It is heated with stirring at 70 °C. A clear yellow solution of [PdCl4]2− was observed and filtered. After that, the ligand
(3,4,3′,4′-tetraaminobiphenyl hydrochloride) (0.0601 g, 1.0 mM equivalent) and the ligand (ethylenediamine) (0.01 g, 1.0 mM) were dissolved, respectively, in 20 mL of distilled H2O. They were then added dropwise to the [PdCl4]2− solution. The pH value was adjusted to 2.3 and 2.6, respectively, by the addition of HCl. Pale yellow and grey precipitate of [Pd(TAB)Cl2] and [Pd(en)Cl2] were formed, respectively. The precipitates [Pd(TAB)Cl2] and [Pd(en)Cl2] were stirred for an additional 30 min at 50 °C and then filtered by filter paper. The precipitates were thoroughly washed with H2O, ethanol, and diethyl ether. The diaqua analogues of [Pd(TAB)Cl2] and [Pd(en)Cl2] complexes were prepared in situ by the addition of slightly less than two mole equivalents of AgNO3. This reaction is proceeded in the dark and left overnight. Then, white precipitate of AgCl that formed was filtered off using a 0.2 μm pore membrane filter. The obtained filtered solution is then completed to 100 mL in a measuring flask and stored. Great care was taken to ensure that the resulting solution was free of Ag+ ion and that the dichloro complexes have been converted completely into the diaqua species.
Synthesis of mixed amino acid complexes [Pd(TAB)(meth)] (3) and [Pd(en)(sar)] (4) was prepared by the addition of 1 mmol equivalent of methionine (W = 0.0237 g) and sarcosine (W = 0.01 g), respectively, then dissolved in 20 mL of distilled water. Then, it was added to [Pd(TAB)Cl2] and [Pd(en)Cl2] to form [Pd(TAB)(meth)] and [Pd(en)(sar)], respectively. The precipitate was stirred for an additional 30 min at 50 °C. [Pd(TAB)(meth)], [Pd(C12H11N4)(C5H11NO2S)], Table 1. Calcd: C, 44.2; H, 4.98; N, 12.9%. Found: C, 43.6; H, 5.01; N, 12.7%. (%) and for [Pd(en)(Sar)], [Pd(C2H4(NH2))2(C3H7NO2)]: Calcd.: C, 20.8; H, 3.5; N, 14.4. Found: C, 20.07; H, 3.01; N, 14.05%. The structure of the synthesized complexes (1)–(4) is shown in Figure 1.
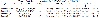
Table 1: Analytical and physical data for [Pd(TAB)(Meth)] and [Pd(en)(Sar)].
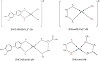
Figure 1: Structural formula of complexes.
Absorption titration experiments were carried out by varying the DNA concentration in the range of (30, 40, 50, 60, 80 μM) as for complex (1) and maintaining the complex concentration constant at (20 μM). As for complex (2), the DNA concentration was in the range of (30, 40, 50, 60, 80 and 160 μM) and a concentration of (20 μM) of complex (2). Upon measuring the absorption spectra, the reference solution was the Tris-buffer solution. Absorbance values were recorded after each successive addition of DNA solution. The sample solution was scanned in the range of 200–600 nm. The absorption data were analyzed for an evaluation of the intrinsic binding constant, Kb, of the complexes with CT-DNA.
Thermal denaturation experiments were carried out by monitoring the change in the absorption of CT-DNA at 260 nm at various temperatures to evaluate the melting temperature (Tm). However, Tm is defined as the temperature at which 50% of double stranded DNA becomes single stranded. Tm was measured in the absence and in the presence of the complexes in Tris-HCl buffer pH 7.1 containing a mixture of 60 μM CT-DNA and 20 μM of the complex. The mixture was stirred continuously, and the temperature was elevated gradually from 30 °C to 90 °C with a reading of absorbance taken every 5 °C. All experiments were made using TB-85-thermobath Shimadzu model UV spectrophotometer equipped with cell-temperature controller. The denaturation temperature (Tm) was taken as the midpoint of the hyperchromic transition. All measurements of Tm were repeated three times and the data presented are the average values.
All the electrochemical measurements were carried out at room temperature in a 15 mL electrolytic cell by using 5 mM Tris-HCl/50 mM NaCl buffer (pH 7.1) as the supporting electrolyte. The GCE surface was freshly polished to a mirror prior to each experiment with 0.05 mm α-Al2O3-paste and then cleaned in water. The working electrode was cleaned after every electrochemical assay. The voltammogram of 15mL of the solution of the complexes, [complex] = 100 μM, was recorded in the absence of DNA. The procedure was then repeated for systems of 15 mL of a mixture containing constant concentration of the complexes ([complex] = 100 μM) and varying the concentration of DNA. For complex (1), 100 μM of the complex was mixed with ([DNA] = 100 and 150 μM) in the molar ratios (1:1 and 1:1.5), respectively. For complex (2), the concentration of the DNA used was ([DNA] = 80 and 100 μM), in the molar ratios (1:0.8 and 1:1).
Ostwald Viscometer was used to perform viscosity titration studies through varying the concentration of the two chelates (1) and (2) at (30, 40, 50, 60, 80) μM with (100 μM) CT-DNA concentration. The flow time of the samples was carefully measured several times and an average value of all measurements was taken at the end. The data is plotted as the binding ratio ([complex]/[DNA]) versus (η/η0)1/3, in which η0 was the viscosity value of free CT-DNA and η was the viscosity value for DNA in the presence of either (1) or (2) [25].
The Pd(II) diaqua complexes (1) and (2) were evaluated for their antibacterial activity against Bacillus subtilis and Staphylococcus aureus (as Gram-positive bacteria) and Escherichia coli and Neisseria gonorrhoeae (as Gramnegative bacteria) using the disc diffusion technique [26] as described in British Pharmacopoeia (2000). Paper discs of
Whatman filter paper (no. 42) of uniform diameter (2 cm) were sterilized in an autoclave. The paper discs, soaked in the desired concentration of the complex solutions, were placed aseptically in the petri dishes containing nutrient agar media (agar 15 g + beef extract 3 g + peptone 5 g) seeded with S. aureus and B. subtilis bacteria separately. The petri dishes were incubated at 37 °C and the inhibition zones
were recorded after 24 h of incubation. The antibacterial activities are calculated as a means of three replicates. The antibacterial activity of a common standard antibiotic Tetracycline was also recorded using the same protocol
as above and at the same concentration and solvent. The antibacterial results of the compounds were compared with the standard and the % activity index for the complexes was calculated by using the formula:
|
|
(1) |
Antitumor activity in vitro was evaluated by using a system based on the tetrazolium salt (MTT). The antitumor activity was tested on two cancer cell lines: the breast cancer (MCF-7) and colon cancer cell line (HCT-116), as well as one normal cell line: the human normal melanocytes (HFB4). Cells were cultured at 37 °C under a humidified atmosphere of 5% CO2 in RPMI 1640 medium supplemented with 10% fetal serum and dispersed in replicate 96-well plates with 1 × 104 cells per well before treatment with the complexes. Various concentrations of the tested complexes (0, 50, 100, 150, 200 and 250 μM) were incubated to the cells with the studied complexes. Monolayer triplicate wells were utilized for each individual dose. After 72 h exposure to the toxins, cell viability was determined by measuring the absorbance at 570 nm with an enzyme-linked immunosorbent assay (ELISA) reader. Each test was performed in triplicate. The relation between surviving fraction and complex concentration is plotted to get the survival curve of each tumor cell line after the specified complex. The IC50 values were derived from the experimental data to provide an estimate of the drug concentration required to block 50% of the desired action. Assays were performed at the National Cancer Institute in Cairo University, Egypt.
The IR spectra of the mixed palladium complexes (3) and (4) are indicated in Table 2. It reveals the presence of the band in the region 3073, 3101 cm−1 characteristics of the (N─H) stretching mode is shifted towards lower frequencies in the spectrum of the palladium complex indicating the involvement of the nitrogen in chelation with the palladium ion [27]. This is further supported by the appearance of a new two-medium intensity band (527, 567 cm−1, respectively) at the region 520–570 cm−1 assignable to (M─N) vibration [28]. The (C⚌O) band of the free amino acid shows two bands at 1610–1660 and 1395–1430 cm−1 region corresponding to the antisymmetric and symmetric (COO−) stretching vibrations, respectively. On complexation, these bands shift to lower and higher wave number, respectively, indicating that the amino acid carboxylate group is involved in complex formation. The symmetric (COO−) stretching band appears at 1427 cm−1 for complex (4) due to the coordination of carboxylic group of amino acid to the metal ion through oxygen. This is further supported by the appearance of (M─O) band in the 454 cm−1 region which confirms the coordination of the amino acid through oxygen [25]. The participation of the NH2 group is confirmed by clarifying the effect of chelation on the in-plane bending, (NH2) vibration. The band appearing at (1253, 1277 cm−1, respectively) in the complexes indicates the participation of the NH2 group in complex formation [29]. Also, a characteristic band due to (NH) appears in the region 3070–3102 cm−1 in complex (4) [30]. The alkyl CH2 group shows characteristic stretching absorption bands in the region (3203, 2765 cm−1, respectively). As for complex (3), the Pd(II) is bonded by 2N atoms from the bidentate ligand and N atom of N-side and S atom from methionine. The Pd(II) atom in complex (4) is bonded to two N atoms from the bi-dentate ligand and one carboxylate O atom and one N atom from sarcosine [31].

Table 2: Tentative assignment of the important infrared bands of [Pd(TAB)(Meth)] and [Pd(en)(Sar)].
One of the most common techniques in DNA-binding studies of metal complexes is electronic absorption spectroscopy. The magnitude of spectral perturbation is evidence for DNA-binding [32]. The absorption titrations were carried out by using a fixed amount of each metal complex (20 μM) with increasing concentrations of DNA in the range of 0–80 μM. The reference solution was the corresponding buffer solution. While measuring the absorption spectra, an equal amount of DNA was added to both the compound solution and the reference solution to eliminate the absorbance of DNA itself. Each sample solution was scanned in the range of 200–600 nm. The absorption spectra of the title complexes with increasing concentrations of CT-DNA are given in Figures 2 and 3, respectively. On increasing the concentration of CT-DNA, the absorption bands of the complexes at 255 nm for complex (1) and at 251 nm for complex (2) were affected, resulting in hyperchromicity and slight red shift. The absorption intensity is increased since the purine and pyrimidine DNA-bases are exposed because of the binding of the complexes to DNA. Metal complexes may bind to DNA with several binding modes, As intercalation or non-intercalation, such as groove binding and binding to the phosphate group [33]. In the present study, it is assumed that the positively charged diaqua complexes (1) and (2) would electrostatically interact with the negatively charged phosphate backbone of the double helix of CT-DNA. This electrostatic binding mode of the two complexes to CT-DNA is also showed by the strong hyperchromism obtained for both complexes suggesting tight binding to CT-DNA and stabilization. The observed strong hyperchromism is most pronounced in the π-π∗ transition, indicating that the complexes are actively associated with the DNA. Those results show agreement with previous studies that discussed that amine groups can rest in the minor groove of GC base pairs [34,35]. As far as the Pd(II) is concerned, it is likely that the ligands facilitate the formation of van der Waal’s contacts within the walls of groove or hydrogen bonds in DNA grooves when interacting with DNA in Tris-HCl buffer. In order to further illustrate the binding strength of the palladium(II) complexes with CT-DNA, the intrinsic binding constant Kb was determined from the spectral titration data. It can be calculated by observing the changes in absorbance at the corresponding λmax with increasing concentrations of CT-DNA, using the following equation [36]:
|
|
(2) |
where [DNA] is the concentration of DNA in base pairs, εf, εa and εb correspond to the extinction coefficient, respectively, for the free diaqua complexes of palladium(II), for each addition of DNA to the palladium(II) complex and for the palladium(II) complex in fully bound form. A plot of [DNA]/(εa − εf) versus [DNA] gives Kb, insets in Figures 2 and 3, as the ratio of the slope to the intercept. From the [DNA]/(εa − εf) versus [DNA] plots, the intrinsic binding constant Kb for complex (1) was 8.36 × 103 M−1 (R2 = 0.87 for six points) and that for complex (2) was 4.25 × 103 M−1 (R2 = 0.99 for eight points). The higher Kb value obtained for complex (1) may plausibly be due to more base pairs available for binding than in complex (1). The calculated Kb values for complexes (1) and (2) are of lower magnitude than that of the traditional intercalator EB (ethidium bromide) (Kb = 1.23 (±0.07) × 105 M−1) [37] and both reveal a strong binding to CT-DNA.

Figure 2: Absorption spectra of complex [Pd(TAB)(H2O)2] (1) in Tris-HCl buffer upon addition of CT-DNA. [complex] = 20 μM, [DNA] = (0) [1], (30) [2], (40) [3], (50) [4], (60) [5], (79.8) [6] μM. Arrow shows the absorbance changing upon the increase of DNA concentration. Inst: plot of [DNA]/(εa − εf) versus [DNA] for the titration of CT-DNA.

Figure 3: Absorption spectra of complex [Pd(en)(H2O)2]2+ (2) in Tris-HCl buffer upon addition of CT-DNA. [complex] = 20 μM, [DNA] = (0) [1], (29.2) [2], (43.6) [3], (59.3) [4], (59.3) [5], (77.8) [6], 160 [7] μM. Arrow shows the absorbance changing upon the increase of DNA concentration. Inst: plot of [DNA]/(εa − εf) versus [DNA] for the titration of CT-DNA with the complex.
The thermal behavior of DNA in the presence of the studied complexes can give additional information about the interaction strength of these complexes with DNA. The double-stranded DNA tends to gradually separate into single strands due to increasing the solution temperature. This dissociation of a duplex nucleic acid into two single strands results in significant hyperchromism around 258 nm. As to identify this transition process, the melting temperature Tm was measured (Tm of the DNA was determined as the transition midpoint). The binding of a ligand to a nucleic acid stimulates its conformational change to increase the denaturation temperatures depending on the strength and mode of its interaction with the nucleic acid. In general, only a small change in the thermal denaturation resulted by the groove binding or the electrostatic attraction along the DNA phosphate backbone. As for intercalation, it leads to a significant rise in thermal denaturation temperature of DNA due to the stabilization of the Watson–Crick base-paired duplex [38,39]. Therefore, the thermal denaturation experiment of DNA provides a suitable tool for detecting binding and evaluating relative binding strengths. The melting curves of CT-DNA in the absence and presence of complexes (1) and (2) are presented in Figure 4. The results obtained indicate that the thermal denaturation temperature of CT-DNA in the absence of complexes (unbound CT-DNA) is 75 °C under our experimental conditions. After adding the complexes, the denaturation temperature increased by nearly 5 °C for each one (ΔTm = 5). Here, ΔTm = Tm − , where Tm and refer to the melting temperature of DNA in the presence and absence of complexes, respectively. The extent of ΔTm is in the interface between the value induced by electrostatic and intercalative binding, and it is relatively lower as compared to those observed for common organic intercalators such as ethidium (13 °C) [40] and some derivatives of porphyrins (≈ 15 °C) [41,42]. This indicates that the two binary Pd(II) complexes most probably bind to CT-DNA by electrostatic and/or groove binding mode, which is in accordance with the results obtained from the absorption titration experiments and from the calculation of Kb.

Figure 4: Melting curves of CT-DNA in Tris-HCl buffer in the absence and presence of complexes, [DNA] = 100 μM, [complex] = 20 μM, showing an increase in Tm due to binding.
Cyclic voltammetry is one of the most important electroanalytical techniques employed for the interaction of metal complexes with biomolecules due to the similarity between various redox chemical and biological processes [43]. It is extremely useful in probing the nature and mode of DNA binding of metal complexes and it provides a useful complement to the above method of investigation such as UV spectra [44]. The redox behavior of the two studied complexes (1) and (2) in the absence and presence of CT-DNA was studied at room temperature within potential range of −1 to 1 V for complex (1) and −1 to 1 V for complex (2) at a scan rate of 200 mV s−1. The results indicate that the two complexes are redox active and show quasi-reversible redox cyclic voltammetric response. The cyclic voltammogram of complex (1) exhibits a quasireversible redox wave with Epc and Epa values of 57 and 176.26 mV, respectively. The ratio of cathodic to anodic peak currents Ipc/Ipa was 0.53. The formal electrode potentials E1/2, ΔEp (difference in cathodic Epc and anodic Epa peak potentials) were 116.5 and 119.2 mV, respectively. At constant temperature, the addition of CT-DNA resulted in the shift in E1/2 = 128 mV and ΔEp = 256 mV (Figure 5), respectively. The ratio of Ipc/Ipa is 0.89 for CT-DNA bound metal complex (1). The shift in potentials and increase in current ratio suggest the binding of complex (1) to CT-DNA [45]. The cyclic voltammogram of complex (2) at scan rate 200 mV s−1 features a quasi-reversible redox wave with E1/2, ΔEp and Ipc/Ipa values of 78.915 mV, 218 mV and 1.129, respectively. Upon the addition of CT-DNA under the same recording conditions, complex (2) experiences a shift in E1/2 (96.3 mV), ΔEp (208 mV) and the ratio of cathodic to anodic peak currents Ipc/Ipa is 0.984 (Figure 6).

Figure 5: Cyclic voltammograms of complex [Pd(TAB)(H2O)2]2+ (1) in Tris-HCl buffer in the absence (a) and presence (b) of CT-DNA [80 μM], (c) CT-DNA [100 μM], V = 200 mV s−1, [complex] = 100 μM; [DNA]: (a) 0, (b) 100, (c) 150 μM.

Figure 6: Cyclic voltammograms of complex [Pd(en)(H2O)2]2+ (2) in Tris-HCl buffer in the absence (a) and presence (b) of CT-DNA [80 μM], (c) CT-DNA [100 μM], V = 200 mV s−1, [complex] = 100 μM; [DNA]: (a) 0, (b) 80, (c) 100 μM.
The mode of the drug vs DNA interaction can be evaluated from the variation in formal potential. In general, it was reported that the positive shift (anodic shift) in formal potential is caused by the intercalation of the cationic drug into the double helical structure of DNA [46], while negative shift is observed for the electrostatic interaction of the cationic drug with the anionic phosphate of DNA backbone [47,48]. As it is obvious, complexes (1) and (2) possess positive peak potential shift indicating (anodic shift) in the CV behavior. Both shifts observed are caused by the addition of DNA which supports our previous assumption groove binding of the positively charged diaqua complexes with the double helix DNA. The effect of the concentration of CT-DNA on potential and current peaks of the complexes (1) and (2) was also studied using a constant concentration of the complex (100 μM) and varying the concentration of DNA.
With complex (1): ([DNA] = 100 and 150 μM) and with complex (2): ([DNA] = 80 and 100 μM), the results showed that the incremental addition of CT-DNA to both complexes causes a diminution of the peak currents due to the variation of the binding state and slowing the mass transfer of the complex after binding to CT-DNA fragments as well as a shift in the E1/2. The decrease of the anodic and cathodic peak currents of the complexes in the presence of DNA is due to the formation of slowly diffusing complex-DNA supramolecular complex, which in turn lowers the concentration of the free complex (mainly responsible for the transfer of current). The shift in the value of the formal potential (ΔE0) can be used to estimate the ration of equilibrium binding constants (KR/K0) according to the model of interaction described by Bard and Carter equation (3) [49]. From this model, one can obtain that
|
|
(3) |
where and are the formal potentials of the bound and free complex forms, and KR and K0 are the corresponding binding constants for the binding of reduction and oxidation species to CT-DNA, respectively. The KR/K0 values for complexes (1) and (2) are 1.56 and 1.967, respectively, suggesting the stronger binding affinity of the reduced form of complexes (1) and (2) compared to the oxidized form. This is further confirmed by the higher cathodic peak potential shift observed for the two complexes, which indicates that the reduced state of complexes (1) and (2) is easier to oxidize in the presence of DNA because its oxidized form is more strongly bound to DNA than its reduced form.
The nature of the binding characteristic of complexes (1) and (2) was further investigated by viscosity measurements on the CT-DNA solution incubated with an increasing concentration of the complexes. The measurements of CT-DNA viscosity are regarded as the least ambiguous and the most critical tests of a binding model in solution in the absence of crystallographic structural data. A plot of relative specific viscosity (η/η0)1/3 versus [complex]/[DNA], where η and η0 are the specific viscosity contribution of DNA in the absence and presence of the complexes, is shown in Figure 7. Cis-platin is well known to kink DNA through covalent binding, shortening the axial length of the double helix [40] and causing a decrease in the relative viscosity of the solution. However, the classical organic intercalators such as ethidium bromide increase the axial length of the DNA and make it more rigid [41,42] based on the classical intercalation model which demands that the DNA helix lengthens as base pairs are separated to accommodate the bound ligand, resulting in an increase in the relative viscosity. As can be seen from Figure 7, a significant increase in the relative specific viscosity of DNA solution was observed with increasing the concentration of both complexes (1) and (2). Thus, we may deduce that the complexes, certainly, are DNA intercalators. From Figure 7, it is also clear that the greater increase in DNA viscosity upon the addition of complex (1) is relatively higher than that obtained upon the addition of complex (2). This also supports the stronger binding ability of complex (1) relative to that of complex (2) which is in agreement with spectrophotometric data.

Figure 7: The effect of increasing the amount of complexes (1) and (2) on the relative viscosity of CT-DNA at 25 ± 0.1 °C, ([DNA] = 20, 40, 60, 80, 100 μM).
It appears interesting to monitor the biological potential of the synthesized diaqua complexes in vivo against different species of bacteria. This is because Pd(II) amine complexes are well known to have enhanced antitumor activity. In testing the antimicrobial activity of these compounds, we used more than one test organism to increase the possibility of discovering antibiotic principles in tested materials. The tested complexes show a notable biological activity against Gram-positive (G+) and Gram-negative (G−) bacteria. The in vitro antimicrobial activity of the complexes was tested against B. subtilis and S. aureus (as Gram-positive bacteria), E. coli and N. gonorrhoeae (as Gram-negative bacteria) and then compared with the standard tetracycline antibacterial agent. The results are listed in Table 3. The results obtained show the following:
(1) [Pd(TAB)(H2O)2] was found to have higher reactivity against the different strains of bacteria more than [Pd(en)(H2O)2] which was found to be moderately active against them.
(2) [Pd(TAB)(H2O)2] exhibits relatively higher reactivity against Gram-negative bacteria than Gram-positive ones.
(3) Both [Pd(TAB)meth] and [Pd(en)sar] exhibit higher reactivity against Gram-positive bacteria than Gram-negative ones, with considerably high reactivity reported for [Pd(TAB)meth] than that of [Pd(en)sar].
(4) With respect to antifungal activity, the four complexes were found to be inactive against A. flavus, but high antifungal activity was reported against the pathogenic yeast C. albicans, especially for complexes (3) and (4).

Table 3: Antimicrobial and antifungal activities of [Pd(TAB)(H2O)2]2+(C1) and [Pd(en)(H2O)2]2+(C2).
It was informed that Gram-negative bacteria have a thin cell wall consisting of few layers of peptidoglycan surrounded by a second lipid membrane containing lipopolysaccharides and lipoproteins. However, Gram-positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids. These differences in cell wall structure can produce differences in antibacterial sensitivity and some antibiotics can kill only Gram-positive bacteria and are ineffective against Gram-negative pathogens [50]. Therefore, the interest in these results is that the two studied complexes (1) and (2) were found to be effective against both Gram-negative and Gram-positive bacteria; furthermore, increasing interests are now directed to the line of antibiotics affecting Gram-negative bacteria since there are certain organisms which have proved difficult to treat and most of them are Gram-negative rods [51,52,53]. It may be assumed that the antibacterial activity of the compounds is related to cell wall structure of the bacteria. This is possible because the cell wall is essential to the existence of bacteria and some antibiotics can kill bacteria by inhibiting a step in the synthesis of peptidoglycan [54,55]. As for the anti-fungal activity, it is suspected that metal complexes increase the delocalization of electrons over the whole chelate ring and enhance the lipophilicity of the complexes. This increased lipophilicity enhances the penetration of the complexes into lipid membranes and blocking of the metal binding sites in the enzymes of microorganisms. These complexes also disturb the respiration process of the cell and thus block the synthesis of the proteins which restricts further growth of the organism.
The cytotoxic effect of complexes may be due to induced oxidative stress which results in cell death by triggering apoptotic processes or necrosis. In this study, the cytotoxic
impact of different concentrations of the two synthesized Pd(II) complexes (1) and (2) in the range of (0, 50, 100, 150, 200 and 250 μM) is being screened successfully against human invasive breast cancer cell line (MCF-7), human colon carcinoma (HCT-116) and normal human melanocyte (HFB4). The cells were exposed to each compound for a span of 48 h so that the complexes would enter the DNA or some other biological target. The IC50 value of the complexes, which indicates the concentration required to make 50% of inhibition in vitro in the lab, was detected from the experimental data. The results are presented in Figures 8(a) and 8(b). The production of MCF-7 and HCT-116 was strongly decreased upon increasing the concentration of complexes. Figure 8(a) shows IC50 value equals 78.1 μM with MCF-7 and 81.9 μM for HCT-116. Figure 8(b) shows that the cytotoxicity of complex (1) against the same tested cancer cells lines is much higher than that observed for complex (2), as shown by the much stronger decrease of the proliferation of MCF-7 and HCT-116 on increasing the complex concentration, with the IC50 value equal to 66.9 μM with MCF-7 and 68.1 μM with HCT-116. Complex (1) shows lower IC50 which reveals its higher tumor inhibitory activity.

Figure 8: Relationship between the concentration of complexes 1 (a) and 2 (b) and the surviving fraction of HCT-116 (human colon cancer cell line), MCF-7 (human breast cancer cell line) and HFB4 (human normal melanocytes).
The study of the interaction of Pd(II) amine complexes with CT-DNA is of special interest for pharmaceutical and biomedical endeavors. The present investigation describes the interaction of two new palladium amine complexes [Pd(TAB)(OH2)2]2+ (1) and [Pd(en)(H2O)2]2+ (2) with CT-DNA. The introduction of amino groups in the Pd(II) coordination sphere would reduce the electron density on the metal center, making it more electrophilic. As a consequence, the acidity of the coordinated water molecule of the diaqua complexes would increase and this would favor the binding of CT-DNA. The binding of the two complexes (1) and (2) was studied by UV-vis spectroscopy and electrochemical studies revealed the strong binding ability of complexes to CT-DNA. The calculated binding strength (Kb) of the two complexes to CT-DNA was found to be of lower magnitude than that of the classical intercalator EB (ethidium bromide) (Kb = 1.23 (±0.07) × 105 M−1) suggesting an electrostatic binding mode. This is also evidenced by small change in the thermal denaturation temperature of DNA (ΔTm) as well as the observed increase in viscosity of DNA after the binding of both complexes. Redox couple of the complexes was assigned as quasi-reversible from their cyclic voltammetric data. The obvious negative peak potential shift by the addition of DNA also supports the electrostatic interaction of the positively charged complexes with the polyanionic DNA.
The antitumor activity of the two complexes (1) and (2) tested on some cancer cell lines as well as on one normal human melanocyte reveals that the cytotoxicity of complex (1) was much higher than that of complex (2); also interesting was the slight effect of complex (1) on the human normal cells, which may consider it a specific target for DNA cancer cells. The antimicrobial tests showed that both complexes exhibited antimicrobial properties, and they were found to be more active against Gram-negative than Gram-positive bacteria. Our findings point to the promising properties of the multidentate N-containing ligands for the design and development of new chemotherapeutic agents.
The authors declare that they have no conflict of interest.
- G. M. Cragg, P. G. Grothaus, and D. J. Newman, Impact of natural products on developing new anti-cancer agents, Chem Rev, 109 (2009), 3012–3043.
- N. Chitrapriya, V. Mahalingam, M. Zeller, and K. Natarajan, Synthesis, characterization, crystal structures and DNA binding studies of nickel(II) hydrazone complexes, Inorganica Chim Acta, 363 (2010), 3685–3693.
- D. B. Hall, R. E. Holmlin, and J. K. Barton, Oxidative DNA damage through long-range electron transfer, Nature, 382 (1996), 731–735.
- R. S. Kumar, S. Arunachalam, V. S. Periasamy, C. P. Preethy, A. Riyasdeen, and M. A. Akbarsha, Surfactant-cobalt(III) complexes: synthesis, critical micelle concentration (CMC) determination, DNA binding, antimicrobial and cytotoxicity studies, J Inorg Biochem, 103 (2009), 117–127.
- J. Reedijk, Why does cisplatin reach guanine-N7 with competing S-donor ligands available in the cell?, Chem Rev, 99 (1999), 2499–2510.
- M. A. Fuertes, C. Alonso, and J. M. Pérez, Biochemical modulation of cisplatin mechanisms of action: Enhancement of antitumor activity and circumvention of drug resistance, Chem Rev, 103 (2003), 645–662.
- M. Nicolini, ed., Platinum and Other Metal Coordination Compounds in Cancer Chemotherapy, vol. 54 of Developments in Oncology, Springer, New York, 1987.
- Q. L. Zhang, J. G. Liu, J. Liu, et al., DNA-binding and photocleavage studies of cobalt(III) mixed-polypyridyl complexes containing 2-(2-chloro-5-nitrophenyl)imidazo [4,5-f][1,10]phenanthroline, J Inorg Biochem, 85 (2001), 291–296.
- A. Divsalar, A. A. Saboury, H. Mansoori-Torshizi, M. I. Moghaddam, F. Ahmad, and G. H. Hakimelahi, Comparative studies on the interaction between bovine β-lacto-globulin type A and B and a new designed Pd(II) complex with anti-tumor activity at different temperatures, J Biomol Struct Dyn, 26 (2009), 587–597.
- R. G. Seddik, F. B. Rashidi, D. S. Salah-Eldin, and A. A. Shoukry, Synthesis, characterization, DNA binding, biological significance, and molecular docking approaches of a palladium(II) complex with ciprofloxacin for more efficient therapy, Chem Biodivers, 21 (2024), e202400415.
- M. D. Coskun, F. Ari, A. Y. Oral, et al., Promising anti-growth effects of palladium(II) saccharinate complex of terpyridine by inducing apoptosis on transformed fibroblasts in vitro, Bioorg Med Chem, 21 (2013), 4698–4705.
- Ž. D. Bugarčić, J. Bogojeski, and R. van Eldik, Kinetics, mechanism and equilibrium studies on the substitution reactions of Pd(II) in reference to Pt(II) complexes with bio-molecules, Coord Chem Rev, 292 (2015), 91–106.
- K. Husain, M. Abid, and A. Azam, Synthesis, characterization
and antiamoebic activity of new indole-3-carboxaldehyde thiosemicarbazones and their Pd(II) complexes, Eur J Med Chem, 42 (2007), 1300–1308.
- C. Icsel, V. T. Yilmaz, F. Ari, E. Ulukaya, and W. T. Harrison, trans-Dichloridopalladium(II) and platinum(II) complexes with 2-(hydroxymethyl)pyridine and 2-(2-hydroxyethyl)pyridine: synthesis, structural characterization, DNA binding and in vitro cytotoxicity studies, Eur J Med Chem, 60 (2013), 386–394.
- A. Divsalar, A. A. Saboury, H. Mansoori-Torshizi, and A. A. Moosavi-Movahedi, Binding properties of a new anti-tumor component (2,2′-bipyridin octylglycinato Pd(II) nitrate) with bovine β-lactoglobulin-A and -B, J Biomol Struct Dyn, 25 (2007), 173–182.
- A. Divsalar, A. A. Saboury, L. Ahadi, et al., Biological evaluation and interaction of a newly designed anti-cancer Pd(II) complex and human serum albumin, J Biomol Struct Dyn, 29 (2011), 283–296.
- A. A. Shoukry and M. S. Mohamed, DNA-binding, spectroscopic and antimicrobial studies of palladium(II) complexes containing 2,2′-bipyridine and 1-phenylpiperazine, Spectrochim Acta A Mol Biomol Spectrosc, 96 (2012), 586–593.
- H. Mansouri-Torshizi, S. Zareian-Jahromi, K. Abdi, and M. Saeidifar, Nonionic but water soluble, [Glycine-Pd-Alanine] and [Glycine-Pd-Valine] complexes. Their synthesis, characterization, antitumor activities and rich DNA/HSA interaction studies, J Biomol Struct Dyn, 37 (2019), 3566–3582.
- F. Mohammadi and H. Mansouri-Torshizi, Five novel palladium(II) complexes of 8-hydroxyquinoline and amino acids with hydrophobic side chains: synthesis, characterization, cytotoxicity, DNA- and BSA-interaction studies, J Biomol Struct Dyn, 38 (2020), 3059–3073.
- D. Wanders, K. Hobson, and X. Ji, Methionine restriction and cancer biology, Nutrients, 12 (2020), 684.
- N. Zhang, Role of methionine on epigenetic modification of DNA methylation and gene expression in animals, Anim Nutr, 4 (2018), 11–16.
- H. Fałtynowicz, M. Daszkiewicz, R. Wysokiński, A. Adach, and M. Cieślak-Golonka, Ni(II) complex with sarcosine derived from in situ generated ligand: structural, spectroscopic, and DFT studies, Struct Chem, 26 (2015), 1555–1563.
- N. S. Sultan, H. K. A. Elhakim, A. A. Shoukry, and F. B. Rashidi, Synthesis, characterization, and biological evaluation of Cu(II) complexes containing triflupromazine with glycine and histidine, Chem Biodivers, 20 (2023), e202300450.
- E. Ishow, A. Gourdon, J.-P. Launay, C. Chiorboli, and F. Scandola, Synthesis, mass spectrometry, and spectroscopic properties of a dinuclear ruthenium complex comprising a 20 Å long fully aromatic bridging ligand, Inorg Chem, 38 (1999), 1504–1510.
- J. Kuncheria and K. K. Aravindakshan, Copper(II) and cobalt(II) complexes of arylazopyrazolones, Synth React Inorg Met-Org Chem, 23 (1993), 1469–1484.
- D. Liu and K. Kwasniewska, An improved agar plate method for rapid assessment of chemical inhibition to microbial populations, Bull Environ Contam Toxicol, 27 (1981), 289–294.
- Z. Abd El-Wahab, M. Mashaly, A. Salman, B. El-Shetary, and A. Faheim, Co(II), Ce(III) and UO2(VI) bis-salicylatothiosemicarbazide complexes: binary and ternary complexes, thermal studies and antimicrobial activity, Spectrochim Acta A Mol Biomol Spectrosc, 60 (2004), 2861–2873.
- M. Shakir, N. Begum, S. Parveen, P. Chingsubam, and S. Tabassum, Synthesis and physico-chemical studies on a 15-membered hexaaza macrocyclic ligand derived from hydrazine and its complexes with Co(II), Ni(II), Cu(II), and Zn(II), Synth React Inorg Met-Org Chem, 34 (2004), 1135–1148.
- G. G. Mohamed, Synthesis, characterization and biological activity of bis(phenylimine) schiff base ligands and their metal complexes, Spectrochim Acta A Mol Biomol Spectrosc, 64 (2006), 188–195.
- E. M. Saad, M. S. El-Shahwai, H. Saleh, and A. A. El-Asmy, Studies on bismuth(III) complexes of ligands containing nitrogen/sulfur and extractive procedure for determination of Bi(III), Transition Met Chem, 32 (2007), 155–162.
- M. S. Mohamed, A. A. Shoukry, and A. G. Ali, Synthesis and structural characterization of ternary Cu (II) complexes of glycine with 2,2′-bipyridine and 2,2′-dipyridylamine. The DNA-binding studies and biological activity, Spectrochim Acta A Mol Biomol Spectrosc, 86 (2012), 562–570.
- P. Zhao, J.-W. Huang, W.-J. Mei, J. He, and L.-N. Ji, DNA binding and photocleavage specificities of a group of tricationic metalloporphyrins, Spectrochim Acta A Mol Biomol Spectrosc,
75 (2010), 1108–1114.
- M. Yodoshi, M. Odoko, and N. Okabe, Structures and DNA-binding and cleavage properties of ternary copper(II) complexes of glycine with phenanthroline, bipyridine, and bipyridylamine, Chem Pharm Bull (Tokyo), 55 (2007), 853–860.
- S. Mylonas, A. Valavanidis, V. Voukouvalidis, and M. Polyssiou, Platinum(II) and palladium(II) complexes with amino acid derivatives. Synthesis, interpretation of IR and 1H NMR spectra and conformational implications, Inorganica Chim Acta, 55 (1981), 125–128.
- N. S. Sultan, A. A. Shoukry, F. B. Rashidi, and H. K. A. Elhakim, Biological applications, in vitro cytotoxicity, cellular uptake, and apoptotic pathway studies induced by ternary Cu (II) complexes involving triflupromazine with biorelevant ligands, Cell Biochem Biophys, 82 (2024), 2651–2671.
- A. M. Pyle, J. P. Rehmann, R. Meshoyrer, C. V. Kumar, N. J. Turro, and J. K. Barton, Mixed-ligand complexes of ruthenium(II): factors governing binding to DNA, J Am Chem Soc, 111 (1989), 3051–3058.
- A. Dimitrakopoulou, C. Dendrinou-Samara, A. A. Pantazaki, M. Alexiou, E. Nordlander, and D. P. Kessissoglou, Synthesis, structure and interactions with DNA of novel tetranuclear, [Mn4(II/II/II/IV)] mixed valence complexes, J Inorg Biochem, 102 (2008), 618–628.
- Mudasir, E. T. Wahyuni, D. H. Tjahjono, N. Yoshioka, and H. Inoue, Spectroscopic studies on the thermodynamic and thermal denaturation of the ct-DNA binding of methylene blue, Spectrochim Acta A Mol Biomol Spectrosc, 77 (2010), 528–534.
- B. Nordén and F. Tjerneld, Binding of inert metal complexes to deoxyribonucleic acid detected by linear dichroism, FEBS Lett, 67 (1976), 368–370.
- M. Cory, D. D. McKee, J. Kagan, D. W. Henry, and J. Allen Miller, Design, synthesis, and DNA binding properties of bifunctional intercalators. Comparison of polymethylene and diphenyl ether chains connecting phenanthridine, J Am Chem Soc, 107 (1985), 2528–2536.
- D. H. Tjahjono, T. Akutsu, N. Yoshioka, and H. Inoue, Cationic porphyrins bearing diazolium rings: synthesis and their interaction with calf thymus DNA, Biochim Biophys Acta, 1472
(1999), 333–343.
- D. H. Tjahjono, S. Mima, T. Akutsu, N. Yoshioka, and H. Inoue, Interaction of metallopyrazoliumylporphyrins with calf thymus DNA, J Inorg Biochem, 85 (2001), 219–228.
- S. Tabassum, I. U. Bhat, and F. Arjmand, Synthesis of new heterometallic macromolecules: their DNA binding, cleavage activity and in vitro model electrochemotherapy study, Spectrochim Acta A Mol Biomol Spectrosc, 74 (2009), 1152–1159.
- Z.-Q. Liu, Y.-T. Li, Z.-Y. Wu, and S.-F. Zhang, [Cu4(H2O)4(dmapox)2(btc)]n · 10nH2O: the first two-dimensional polymeric copper(II) complex with bridging μ-trans-oxamidate and μ4-1,2,4,5-benzentetracarboxylato ligands: Synthesis, crystal structure and DNA binding studies, Inorganica Chim Acta, 362 (2009), 71–77.
- A. J. Bard and L. R. Faulkner, Electrochemical Methods: Fundamentals and Applications, John Wiley & Sons, New York, 2nd ed., 2001.
- M. Aslanoglu, Electrochemical and spectroscopic studies of the interaction of proflavine with DNA, Anal Sci, 22 (2006), 439–443.
- N. Li, Y. Ma, C. Yang, L. Guo, and X. Yang, Interaction of anticancer drug mitoxantrone with DNA analyzed by electrochemical and spectroscopic methods, Biophys Chem, 116 (2005), 199–205.
- A. Shah, M. Zaheer, R. Qureshi, Z. Akhter, and M. F. Nazar, Voltammetric and spectroscopic investigations of 4-nitrophenylferrocene interacting with DNA, Spectrochim Acta A Mol Biomol Spectrosc, 75 (2010), 1082–1087.
- M. T. Carter and A. J. Bard, Voltammetric studies of the interaction of tris(1,10-phenanthroline)cobalt(III) with DNA, J Am Chem Soc, 109 (1987), 7528–7530.
- A. L. Koch, Bacterial wall as target for attack: Past, present, and future research, Clin Microbiol Rev, 16 (2003), 673–687.
- H. Nikaido and T. Nakae, The outer membrane of Gram-negative bacteria, Adv Microb Physiol, 20 (1979), 163–250.
- S. P. Datta, Resistance of Pseudomonas aeruginosa, John Wiley & Sons, London, 1975: edited by M. R. W. Brown, Biochem Educ, 4 (1976), 62–62.
- M. A. González, Synthetic derivatives of aromatic abietane diterpenoids and their biological activities, Eur J Med Chem, 87 (2014), 834–842.
- A. A. El-Sherif, Synthesis, spectroscopic characterization and biological activity on newly synthesized copper(II) and nickel(II) complexes incorporating bidentate oxygen-nitrogen hydrazone ligands, Inorganica Chim Acta, 362 (2009), 4991–5000.
- S. Mylonas, A. Valavanidis, V. Voukouvalidis, and M. Polyssiou, Synthesis of S-2-aminoethyl-L-cysteine and S-2-aminoethyl-D,L-penicillamine complexes with Pt(II) and Pd(II). interpretation of IR and 1H NMR spectra and conformational implications, Inorganica Chim Acta, 66 (1982), 25–28.
Copyright © 2024 Radwa Hatem et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.